|
Page History
Choosing Genes
caIntegrator provides three methods whereby you can To obtain gene names for a gene expression search or analysis, use one of the following three methods described in this section: bioDBnet, Gene List or CGAP.
bioDBnetcaBIO – This link searches caBIO, then bioDBnet for gene IDs, symbols or genes within pathways. Then caIntegrator pulls identified genes into caIntegrator the application for analysis.Anchor bioDbNetSearch bioDbNetSearch - Click the caBIO icon () bioDBnet.
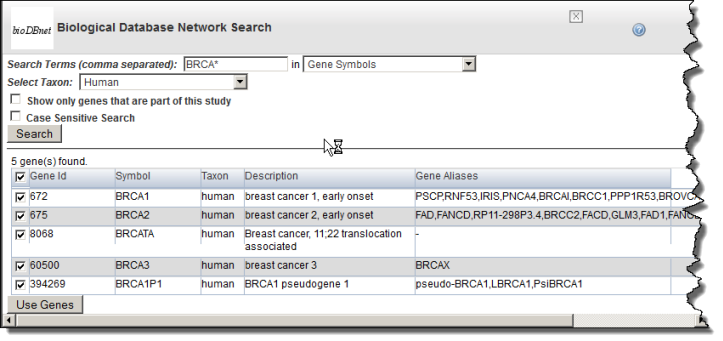
- Enter Search Terms. Note that caIntegrator can perform a search on a partial HUGO symbol. For example, as search using ACH * would find matches with 'achalasia' and 'arachidonate'.
- Select if you want to search in Gene Keywords IDs, Gene Symbols, Gene Alias, Database Cross Reference Identifier or Aliases, Pathways (from the drop-down list), or Search Pathways for Genes.
- Gene Keywords searches the description field in caBIO; the result displays in the Full Name ColumnIDs searches the exact gene ID(s) you enter.
- Gene Symbols searches only the Unigene and HUGO gene symbols in caBIObioDBnet.
- Gene AliasAliases searches for one or more gene symbols which are synonymous for the current gene symbol.
- Database Cross Reference Identifier searches for the symbol for this gene as it appears in other databases.
- Pathways searches only the pathway names in caBIO. Note that searching in Pathways is a two step process. First, the initial Pathway search produces search results which are pathways. Second, from the pathway search results screen, you must select pathways of interest, then click Search Pathways for Genes to obtain a list of genes related to the selected pathways.bioDBnet.
- Search Pathways for Genes searches for pathways containing gene(s) you specify for the search.
- Select Show only genes that are part of this (caIntegrator) study or Case Sensitive Search if either of these criteria are to be applied to the search. (By default, the search is case insensitive.)Select the Any or All choice to determine how your search terms will be matched. Any finds any match for any search term you entered. All finds only results that match all of the search terms.
- Choose the Taxon from the drop-down list and click Search. (The Taxon criterion defaults to Human.) The search results display , as shown in the following figure. on the same page below the search criteria. The following figure shows search criteria and a few of the listed search results.
- In the search results, use the check boxes to identify the genes whose symbols you want to use in the gene expression analysis.
- Click Use Genes at the bottom of the page. This pulls the checked genes into the Gene Symbol text box on the Criteria tab, shown in the following figure. . The following figure reveals some of the genes pulled into the Gene Symbol text box.
- Gene List– This link locates gene lists saved in caIntegrator.
- Click the Genes List icon () to open a Gene List Picker dialog. For more information, see on page 69 Creating a Gene or Subject List.
If a GISTIC analysis has been run, you may see the following options:- GISTIC Amplified genes is a list of gene symbols in which the corresponding regions of the genome are significantly amplified.
- GISTIC Deleted genes is a list of gene symbols in which the corresponding regions of the genome are significantly deleted.
- In the drop-down menu that lists previously saved gene lists, select a gene list. In the list that appears, use the check boxes to identify the genes whose symbols you want to use in the gene expression analysis.
- Click Use Genes at the bottom of the dialog. This pulls the checked genes into the Search Criteria tab.
- Click the Genes List icon () to open a Gene List Picker dialog. For more information, see on page 69 Creating a Gene or Subject List.
- CGAP – Use this directory to identify genes. Before clicking this link the CGAP icon () you must enter gene symbols in the text box. This link does not pull anything into caIntegrator but does provide information about the gene(s) whose names you entered.