|
Page History
caMicroscope is a tool to view, annotate, and analyze whole-slide, biomedical images. It consists of a set of web services to manage manages digital pathology images, associated clinical and imaging metadata, and human/machine generated annotations and markups. It also allows you to view and analyze nuclear segmentations of images. caMicroscope hosts whole-slide, cancer tissue images for review by pathologists.FeatureScape provides interactive representations of feature landscapes. Use FeatureScape to explore slide-level imaging features generated by analysis of whole slide tissue images to QuIP.
This guide explains how to do the following tasks in caMicroscope:
| Table of Contents |
|---|
Using caMicroscope
caMicroscope allows you to view whole-slide images and nuclear segmentations.
Opening caMicroscope
- Use the Chrome browser to navigate to https://wolf.cci.emory.edu/vtr_pending.
The Sign In to caMicroscope.The Login page appears.
SEER VTR caMicroscope Login Page Use your Google credentials to sign in to caMicroscope.
The caMicroscope site appears.
caMicroscope Home Page- Select one of the image databases Select which database of images you want to view by clicking either the large slide image or the the More button button. Databases Options include the Breast Cancer Genomic Pilot, Virtual Tissue Repository (VTR) Pending Slides, and and Pancreatic Ductal Adenocarcinoma (PDAC).
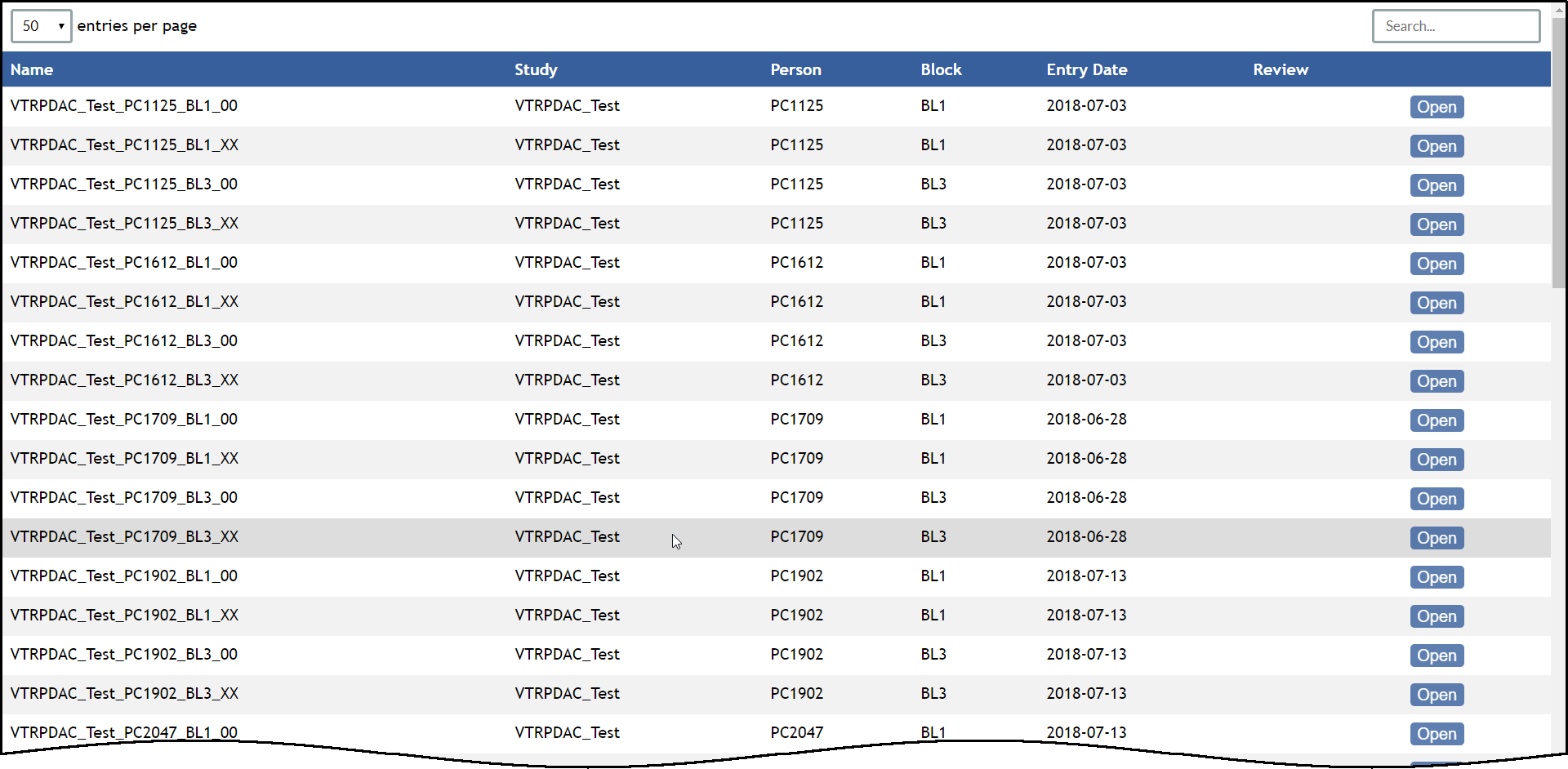
A table appears that lists all whole-slide images for the selected database that you are authorized to view.
caMicroscope Data Table for the PDAC Database In the table, click the the Open button button for any row.
The slide opens in caMicroscope.Note To select a different image, click the Back button in the caMicroscope toolbar or the back arrow in your browser to return to the table.
Using the caMicroscope Toolbar
Use the toolbar buttons to manipulate the slide.
...
Tool
...
Name
...
Purpose
...
Home
...
Return to the caMicroscope home page.
Zooming In and Out
To zoom in on the image, click the main content window with your mouse. Each time you click, the view zooms in further, the scale updates, and the bounding box in the inset window changes to show how much of the total image is in the main content window. When you are zoomed in very far, the bounding box may not be visible. Zooming out reverses this process.
To Zoom In: Click in the main window.
To Zoom Out: Press and hold the Shift key while you click on the main window.
Zoomed in:
Zoomed out:
...
Panning the Slide
Panning allows you to move the image inside the window.
- Select an image and view it in caMicroscope. See Opening caMicroscope for more information.
- In the inset window, note the red bounding box. This bounding box is your view of the current image in the main content window.
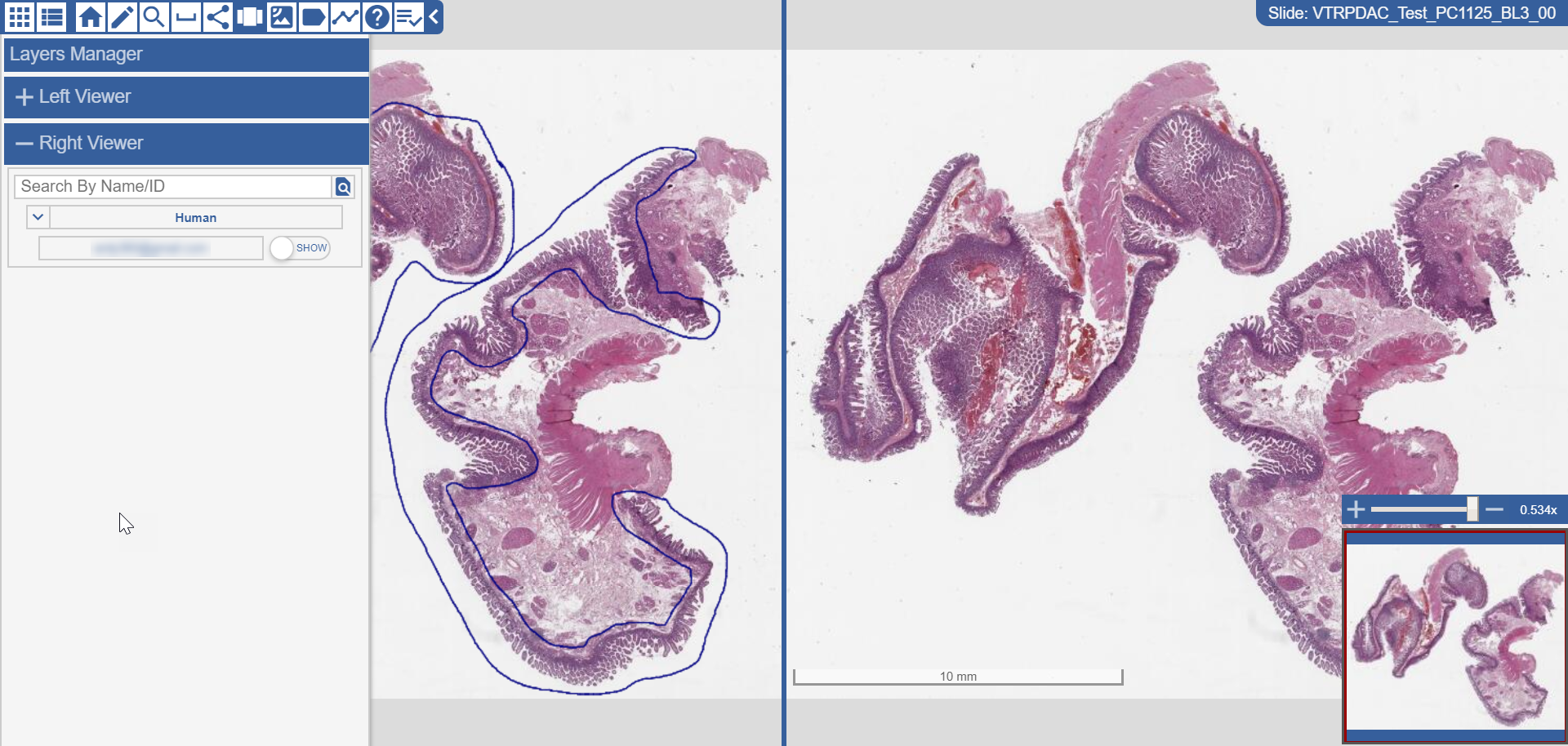
caMicroscope Main Content Window and Inset Window - Drag Click your mouse to control the bounding box and drag it up, down, left, and right to see different parts of along the main content window.
The red bounding box moves to reflect your current position.
Zooming In and Out
The inset window has a zoom slider above it.
- To zoom in on the image, move the slider towards the plus sign.
- To zoom out, move the slider in the inset window towards the minus sign.
As the slider moves, the main content window zooms in or out and the red bounding box in the inset window changes to show how much of the total image is in the main content window.
Using the Lymphocyte & Plasma Cell Annotation App
From caMicroscope you can launch the Lymphocyte & Plasma Cell Annotation App by clicking on the caMicroscope toolbar. This app allows you to view and modify annotations related to lymphocyte and plasma cells.
The following tools are available on the toolbar.
Inset Window with Red Bounding Box and Zoom Slider
Using the caMicroscope Tools
The toolbar is in the top-left of the main content window. Use the toolbar buttons to manipulate the slide. To close any toolbar button, click the same button again or a new button.
caMicroscope Toolbar
Tool | Name | Purpose | |||||||||||||||||
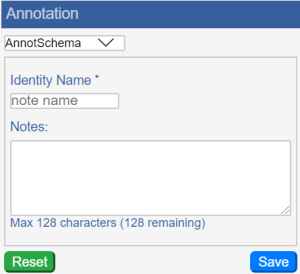
Annotations | Opens the Annotation panel, where you can select which annotation set to view, name that annotation set, add optional notes about the annotation set, save the annotation set, and reset the panel to its original state.
| ||||||||||||||||||
Layer Manager | Opens the Layers Manager panel, where you can select which layers to view. | ||||||||||||||||||
Home | Return to the data table so that you can open another slide. | ||||||||||||||||||
Draw | Home | Return to the caMicroscope home page | caMicroscope | Return to the caMicroscope application | Filter Markups | View only those markups associated with selected algorithms. For more information, see page 8. | Show/Hide Markups | View the image with or without markups. | Decrease Opacity | Decrease the opacity of the selected markup(s), making it/them more transparent. | Increase Opacity | Increase the opacity of the selected markup(s), making it/them less transparent. | Show Weight Panel | This panel allows you to adjust the automatic lymphocyte prediction results for this slide, with the options described below.
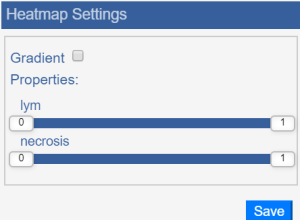
Adjust the sensitivity of lymphocyte prediction. Choose a higher volume for more predicted lymphocyte regions.
Adjust the specificity of necrosis prediction. Choose a higher volume for more predicted lymphocyte regions.
Show lymphocyte prediction without necrosis filtering.
Show necrosis prediction.
Show lymphocyte prediction with necrosis filtering (recommended). | Free Line Markup | Draw thin lines, thick lines, or polygons on the image. | Switch User | If you are a super user, click this button to review and change other people’s annotations. |
Using the Segment Curation App
Use the Segment Curation App to curate segmentation results.
The Segment Curation App has the following tools.
...
Tool
...
Name
...
Purpose
...
Home
...
Return to the caMicroscope home page.
...
Free Line Markup
...
Draw thin lines, thick lines, or polygons on the image.
...
Save ViewPort
...
For a selected algorithm, save the ViewPort.
...
Save Rectangle and Delete Annotations in This Area
...
For an area based on a selected algorithm, save the rectangle and delete the annotations.
...
Filter Markups
...
View only those markups associated with selected algorithms. For more information, see page 8.
...
Generate Composite Dataset
...
ColorMap
Filtering Markups
You can filter your view of the markups on the images in caMicroscope by an area based on one or more selected algorithms. The algorithms available in caMicroscope have been run against the full set of images.
To filter markups
- Click the Filter Markups button.
The Select Algorithm box appears. - Click the box to the left of each algorithm you want to select.
caMicroscope runs the algorithm(s) against each image in the dataset. This can take some time. The markups appear in the color noted in the Select Algorithm box. You may have to zoom in and pan the image to see all markups.
Analyzing Images
To analyze images
...
To maintain the integrity of measurements, avoid drawing shapes that overlap or intersect one another. | ||
| Magnifier | The Magnifier works like a magnifying glass and allows you to see the slide at normal magnification (1.0), low magnification (0.5), or high magnification (2.0). Click a magnification level and place the bounding box on the area of the slide you want to magnify. | |
| Measurement | Drag this tool on the slide to learn the measurement in micrometers.
| |
| Share View | Opens a window with a URL to the current presentation state of the slide including the magnification level, layers that are currently open, and your position on the image. | |
| Side by Side Viewer | Shows the Layer Manager panel, the left and right layers, and inset window. For the right and left layer, select which layer you want to view.
| |
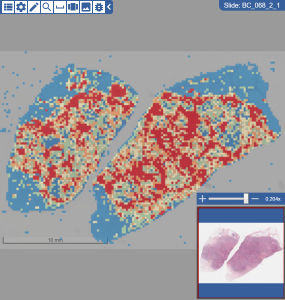
| Heatmap | For a slide with heatmap data, opens the choices of heatmaps available, as well as ways of displaying the heatmaps. The gradient shows all of the values on the selected spectrum for the field you selected. Choices of heatmaps: HeatMap displaying a gradient for lym: Slides with heatmaps have three custom tools in the toolbar.
| |
| Labeling | Use this tool to draw a circle or rectangle around a tumor region, measure an area on the slide, download labels, and submit a bug report. The Labeling tool has its own toolbar with tools in the following order from left to right: return to the previous slide, place a square on the slide, place a circle on the slide, measure an area, download labels, and submit a bug report. Click the left arrow at the far right of the toolbar to hide it, then click the right arrow to show it.
| |
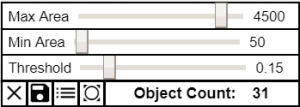
| Segment | This tool allows you to display, count, and export nuclear segmentations on the image. Clicking this tool opens the following custom toolbar. Segment Toobar
| |
| Help | Opens this guide. | |
| Reviewed | Click this button after reviewing a slide to change its review status, then confirm the change. A green checkmark appears in the data table for this slide after you have marked it as reviewed. |
Viewing Scatterplots
To the right of the feature correlation plot is a scatterplot based on the computed feature values. The scatterplot shows how objects are distributed. Zoom in by selecting a region to look at the distribution in more detail.
When you move your cursor over the scatterplot, you see tools you can use in it. Hover your mouse over a tool to view the tool name.
Selecting a Region on the Scatterplot
Select a region on the scatterplot to zoom in to a subset of segmented objects.
To select a region:
- Move your cursor over the scatterplot to see the crosshairs.
- Click an area of interest and drag your cursor to create a rectangle.
- Release the mouse. The scatterplot zooms in to the selection.
You can now view nuclear mugshots or download data from the selected region. To return to the original scatterplot, click the selected pixel in the heat map again.
Viewing Nuclear Mugshots
Nuclear mugshots are screenshots of segmented nuclei in the set of features you selected in the heat map. Only a sampling of the millions of images has associated mugshots.
To view nuclear mugshots:
- Select an area of interest on the scatterplot.
- Click Nuclear mugshots from selected region.
The nuclear segmentation viewer opens, showing tiles of slides that match the range you selected. - Click any tile to open it in caMicroscope and view it in the context of the whole slide image.
NOTE: Make sure you are logged into caMicroscope before you select a tile.
Downloading Data from a Scatterplot
To download data from a scatterplot
- Select an area of interest on the scatterplot.
- Click .
The FeatureScape Data Download page appears. - Review the information on the page to confirm the relative time it will take to download your selection.
- Enter the maximum number of features to download and then click .
The Data Download page refreshes with a list of variables represented in the download. You can select some or all of them and then click .
Alternatively, click Save (partial) File as CSV. This skips the step of selecting individual variables.
...