Page History
| Scrollbar | ||
|---|---|---|
|
...
| Page info | ||||
|---|---|---|---|---|
|
This chapter describes search results that caIntegrator returns after queries.
...
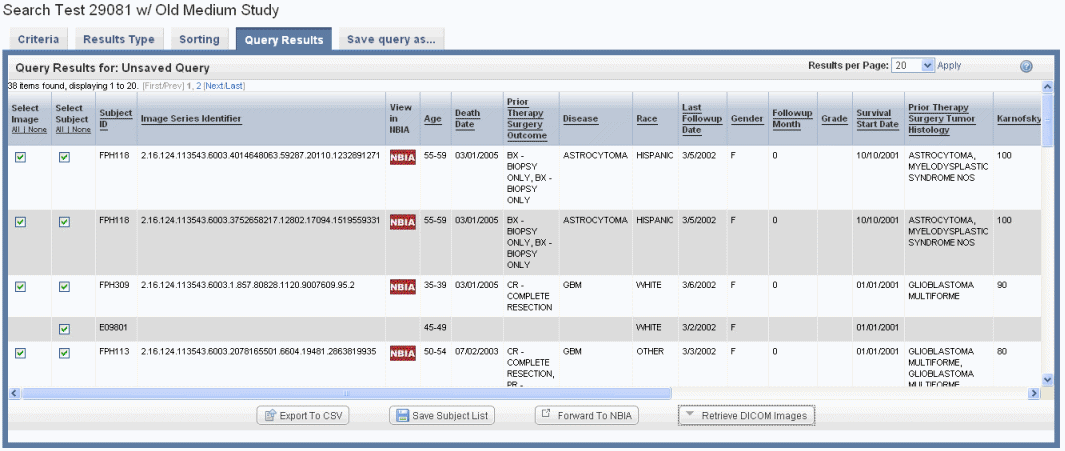
If you run the search before configuring column and sort display parameters, only the [subject] ID that meet the criteria and a column allowing you to select each row appear on the table, as shown in the following figure.
You can add details for one or more subjects by configuring them on the Results Type tab. Annotations listed there are the column headers in the CSV file(s) that were uploaded to the study. For information about using the Results Type tab, see Results Type Tab.
...
For Gene criteria, caIntegrator finds and summarizes at the gene level all reporters that match criteria for the gene you defined on the Criteria tab. When displaying results at the gene level, the mean is displayed if there are two probes, the median if there is an odd number of probes greater than two and the mean of the two median probes if there is an even number of probes greater than two. Next to each gene symbol, caIntegrator displays an icon () which you can click to open the Cancer Genome Anatomy Project (CGAP) showing data for the gene. Gene symbols and icons are identified in the following figure.
If you have selected Gene Expression on the Results Type tab, then the column headers are a clickable label which sorts the entire table on that column. If you selected Reporter ID on the Results Type tab, the Reporter ID is clickable (and the gene is not clickable).
...
Gene reporter type display
Reporter ID reporter type display
...
- Click the Create New List link in the left sidebar. This opens the Manage List page, shown in the following figure.
- Enter a name for the gene or subject list.
- Enter a description (optional).
- Select Make Visible to Others if you want the list to be visible to anyone who views the study. This selection places the list in the Global List folder in the left sidebar under Saved Lists. In any box where you can select lists, the term 'Global' will identify any list so identified when the list is created.
- Select the List Type, Subject or Gene.
- If you select Subject, enter the Subject IDs in the text box that appears. Proceed with step 7.
- If you select Gene, proceed with step 6.
- For Gene Symbol, caIntegrator provides three methods whereby you can obtain gene symbols for creating a gene list. For more information, see Choosing Genes.
- If you so choose, you can upload a gene or subject ID list. For the Upload File field, click the Browse button to navigate to a .csv file made up of gene symbols. caIntegrator converts the comma-separated content to a gene list.
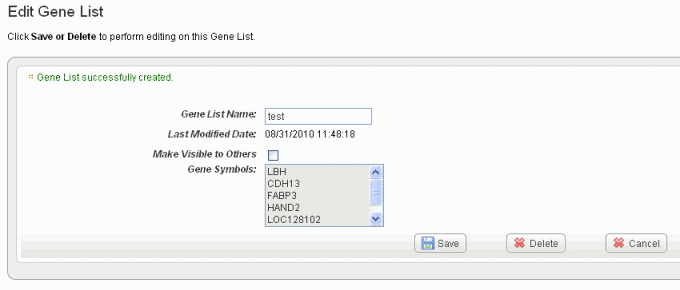
- Click Create List at the bottom of the page. caIntegrator now opens the Edit [Subject or Gene] List page which shows the name and symbols of the newest gene list, shown in the following figure.
See Editing a Gene or Subject List for information about the edit feature.
...
- Click on any of the list names or the list icon () to rerun the query from which the gene or subject list was first created. If the list is a gene list, in the query results, you can click on the gene information icon () to open the Cancer Genome Anatomy Project (CGAP) showing metadata for the gene.
- Click the edit icon () to open an Edit Gene/Subject List dialog box. On this page you can review the list of gene symbols or subject IDs included in the list shown in the following figure.
- In the Edit [List Type] dialog box, you can perform the following tasks:
- To rename the list in the [List Type] List Name text box, enter the new list name.
- You can change the visibility of the list in the appropriate check box.
- To delete the list, click the Delete button.
- Click Save to save your changes or Cancel to leave the page without making changes.
...
If your annotation choice on the Results Type page identifies annotations such as tumor size or tumor location, the search results display image series subsets that have those annotations, or any annotations you check on the Results Type page. The check boxes work in conjunction with buttons at the bottom of the results page, shown in the following figure. By expanding display parameters, you can view complete details for image search results.
You can add more details for images by configuring image annotations on the Results Type tab. Annotations listed there are the column headers in the image series CSV file(s) that were uploaded to the study. Examples of image details include the following:
...
On the caIntegrator imaging data Search Results page, you can click the Retrieve DICOM Images button which is linked to results you have selected by row. caIntegrator retrieves the corresponding image(s) from NBIA through the grid. NBIA organizes the download file by patient ID, StudyInstance UID, and ImageSeries UID, and compresses it into a zip file. When caIntegrator notifies you that the file is retrieved, the DICOM Retrieval page indicates whether the retrieved files are Study Instance UIDs or Image Series UIDs, shown in the following figure.
Click the Download DICOM link to download and save the file. caIntegrator unzips the file and displays the list of images in the file. To open the DICOM images, you must have a DICOM image viewer application installed on your computer. For more information about one such viewer, see http://www.e-dicom.com/viewers.php
| Multiexcerpt include | ||||||
|---|---|---|---|---|---|---|
|
...
| Info | ||
|---|---|---|
| ||
You will not see the Export option when gene expression data displays as query results. | ||
| Wiki Markup |
| Scrollbar | ||
|---|---|---|
|