|
Page History
- Add data values to Data and Conditions.
- To import a file of data values
- Save the spreadsheet of data values to a csv (comma-separated value) file.
- Click Import csv and select and follow the prompts to add the data file to the Findings Info.
- The columns and data are added to Data and Conditions.
- To add the data values manually
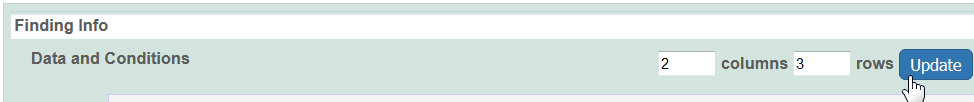
- Specify the number of columns and rows for the matrix, and click Update.
- Add the data values to the rows.
- Specify the number of columns and rows for the matrix, and click Update.
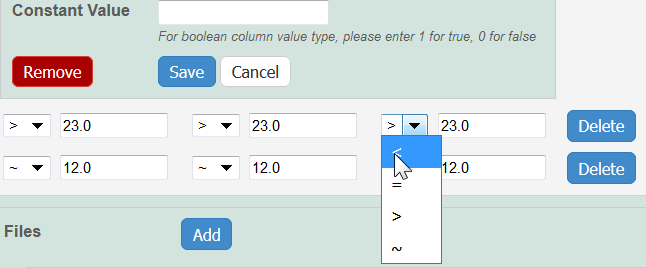
Info Whether you imported or added information manually, you can preface each data value with one of the following: Maintain the default, equal to (=), or select greater than (>), less than (<), or infinity (approximate).
- To import a file of data values
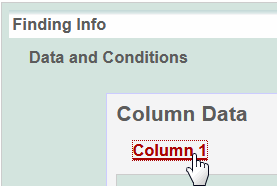
- To define a column, click an underlined column heading.
The Column Definition panel displays.
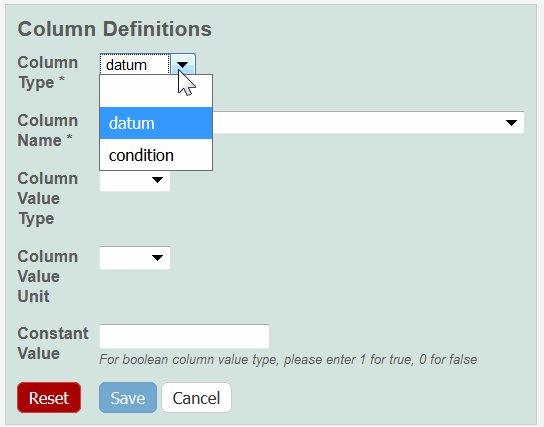
Select a Column Type, Datum or Condition.
Select a Column Name or select other and add a new one.
Info title Column Notes You can add up to three cell viability Column Names, including cell viability, cell viability B, and cell viability C. You can further identify the column with the Column Value Type. For Column Type, Datum, the following characterization(s) display customized Column Name options.
- For the matrix size, enter numbers for rows and columns and click the Update button.
The window now displays column [#] headings that you can name for your matrix. - Click an underlined column heading. Available options can be specific for the type of characteristic you are defining. As described below, select an option or if there are none or if you prefer, select [Other] to name the column yourself.
- Column Type – You must select either Datum or Condition. In the cases noted below, based on your Characterization selection at the top of the Characerization page, customized options may display for the following fields.
- Column Name and Column Property – Select or enter a column name.
If you selected Datum for Column Type (or if appropriate, Condition), the following characterizations display customized name fields. In some cases, based on your selection at the top of the characterization page, customized options may display for some fields.
For a physico-chemical characterization, use the criteria in the first table.
For an in vitro characterization, use the criterion in the second table.
For an in vivo characterization, click [Other] to name the column yourselfColumn Type "Datum" is selected with this characterization
Autopopulated Column Name Option[s]
Molecular Weight
Purity
Relaxivity
Characterization Type Column Type and Column Name Option(s) Physico-Chemical - Molecular – Molecular Weight
- Purity – % purity for sample
- Relaxivity –
- R1, R2, T1, T2
- Size
- – PD1, Peak N , RMS size, Z Average
- Surface
surface area,- – charge,
- zeta potential
Column Type "Datum" is selected with this characterization
In Vitro Enzyme Induction
– % of Control
In Vivo Click Other to name the column yourself. For Column Type, Condition, all characterizations provide the Column Name options
If you selected Condition for Column Type, for all characterizations, Column Name displays the options listedin the left column of the following table.
When you make a name selection, Condition Propertiesoptions may correspond to the name you selected; those are shownThe Column Name autopopulates the Condition Property options in the right column.
Select one of the column properties, where appropriate."Condition" is selected; Column Name is autopopulated with these optionsColumn Type for Column Name, Condition Autopopulates Column Name Column Name Autopopulates
Condition PropertyN/A
media type, serum percentage
bandwidth, frequency, time, wavelength
N/A
lyophilized, time
time
N/A
N/A
lyophilized, time
ion concentration, ionic strength, molecular formula, osmolality, serum percentage, with serum
number of pulses, pulse duration
N/A
To further identify a column, select a Column Value Type
–.
Select one of the value types listed in the Value Type drop-down listInfo Once the column information is saved, the Column Type is shown in parentheses after the Column Name, such as cell viability (mean) . - Select a Column Value Unit – Select or enter the appropriate value unit.
- Constant Value? – Enter a constant value, if appropriate. , or select other and add one.
If you want the same value to fill all rows in a column, add a Constant Value.
Info title For Column Value Type, boolean For Column Value Type, boolean, enter a Constant Value of 1 for true and 0 for false. Click Save, and the column(s) are updated.
Click Set Column Order to indicateInfo If needed, click Set Column Order to change
you have definedthe order of the column headings in the matrix
.
- Click Save to confirm the your matrix design choices. After you do so, the names you selected show as column headings. Click Remove to delete the column, or Cancel to close the window.Click Save on the Finding page, as well in the Finding section.