|
Page History
...
Step | Action | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
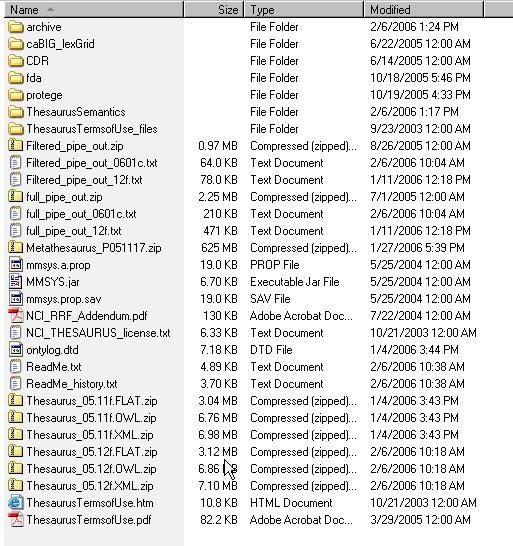
1 | Using a web or ftp client go to the URL: ftp://ftp1.nci.nih.gov/pub/cacore/EVS/ | |||||||||||
2 | Select the version of NCI Thesaurus OWL you wish to download. Save the file to a directory on your machine. | |||||||||||
3 | Extract the OWL file from the zip download and save in a directory on your machine. This directory will be referred to as NCI_THESAURUS_DIRECTORY | |||||||||||
4 | Using the LexEVS utilities load the NCI Thesaurus:
For Windows installation use the following command:
For Linux installation use the following command:
|
Example output from load of NCI Thesaurus 05.12f
| Code Block |
|---|
…
[LexBIG] Processing TOP Node... Retired_Kind
[LexBIG] Clearing target of NCI_Thesaurus...
[LexBIG] Writing NCI_Thesaurus to target...
[LexBIG] Finished loading DB - loading transitive expansion table
[LexBIG] ComputeTransitive - Processing Anatomic_Structure_Has_Location
[LexBIG] ComputeTransitive - Processing Anatomic_Structure_is_Physical_Part_of
[LexBIG] ComputeTransitive - Processing Biological_Process_Has_Initiator_Process
[LexBIG] ComputeTransitive - Processing Biological_Process_Has_Result_Biological_Process
[LexBIG] ComputeTransitive - Processing Biological_Process_Is_Part_of_Process
[LexBIG] ComputeTransitive - Processing Conceptual_Part_Of
[LexBIG] ComputeTransitive - Processing Disease_Excludes_Finding
[LexBIG] ComputeTransitive - Processing Disease_Has_Associated_Disease
[LexBIG] ComputeTransitive - Processing Disease_Has_Finding
[LexBIG] ComputeTransitive - Processing Disease_May_Have_Associated_Disease
[LexBIG] ComputeTransitive - Processing Disease_May_Have_Finding
[LexBIG] ComputeTransitive - Processing Gene_Product_Has_Biochemical_Function
[LexBIG] ComputeTransitive - Processing Gene_Product_Has_Chemical_Classification
[LexBIG] ComputeTransitive - Processing Gene_Product_is_Physical_Part_of
[LexBIG] ComputeTransitive - Processing hasSubtype
[LexBIG] Finished building transitive expansion - building index
[LexBIG] Getting a results from sql (a page if using mysql)
[LexBIG] Indexed 0 concepts.
[LexBIG] Indexed 5000 concepts.
[LexBIG] Indexed 10000 concepts.
[LexBIG] Indexed 15000 concepts.
[LexBIG] Indexed 20000 concepts.
[LexBIG] Indexed 25000 concepts.
[LexBIG] Indexed 30000 concepts.
[LexBIG] Indexed 35000 concepts.
[LexBIG] Indexed 40000 concepts.
[LexBIG] Indexed 45000 concepts.
[LexBIG] Indexed 46000 concepts.
[LexBIG] Getting a results from sql (a page if using mysql)
[LexBIG] Closing Indexes Mon, 27 Feb 2006 01:44:22
[LexBIG] Finished indexing
|
...
Step | Action | ||||||
|---|---|---|---|---|---|---|---|
1 | Using a web or ftp client go to the URL: ftp://ftp1.nci.nih.gov/pub/cacore/EVS/ | ||||||
2 | Select the version of NCI Metathesaurus RRF you wish to download. Save the file to a directory on your machine. | ||||||
3 | Extract the RRF files from the zip download and save in a directory on your machine. This directory will be referred to as NCI_METATHESAURUS_DIRECTORY.
| ||||||
4 | Using the LexEVS utilities load the NCI Thesaurus:
For Windows installation use the following command:
For Linux installation use the following command:
|
...
Step | Action | ||||||
|---|---|---|---|---|---|---|---|
1 | Using the LexEVS utilities load the NCI Thesaurus:
For Windows installation use the following command:
For Linux installation use the following command:
|
...
Step | Action | ||||||
|---|---|---|---|---|---|---|---|
1 | Using a web or ftp client go to the URL: ftp://ftp1.nci.nih.gov/pub/cacore/EVS/ | ||||||
2 | Select the version of NCI History you wish to download. Save the file to a directory on your machine. Select the VersionFile download to the same directory as the history file. | ||||||
3 | Extract the History files from the zip download and save in a directory on your machine. This directory will be referred to as NCI_HISTORY_DIRECTORY | ||||||
4 | Using the LexEVS utilities load the NCI Thesaurus:
For Windows installation use the following command:
For Linux installation use the following command:
|
...