|
Page History
| Scrollbar | ||
|---|---|---|
|
| Page info | ||||
|---|---|---|---|---|
|
| Panel | ||||
|---|---|---|---|---|
| ||||
|
FORMAT THIS PAGE AND THE CHILD PAGES <?xml version="1.0" encoding="utf-8"?>
<html>
...
This document is intended for LexEVS developers.
Document
...
Sections
Related
...
Documents
- Programmer's Guide for information about using the LexEVS core services and the APIs.
Service
...
Integration with caBIG:
...
Diagrams
LexEVS software architecture and implementation is designed to facilitate flexibility and future expansion. The following diagrams are intended to aid the understanding of LexEVS service integration in context of the larger caBIG® caBIG universe and specific deployment scenarios:
...
...
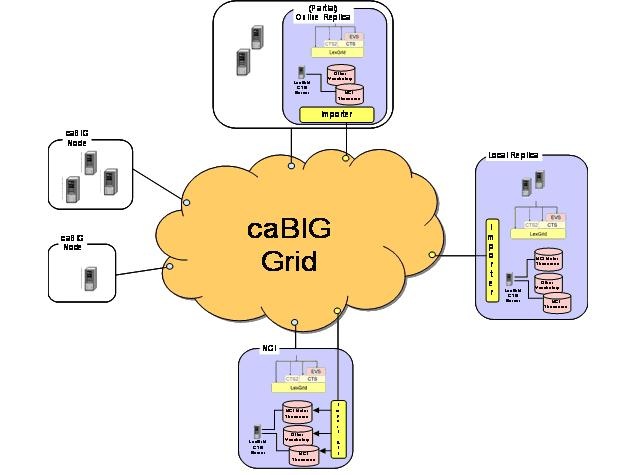
The following diagram depicts the LexBIG vision. Individual Cancer Centers will be able to use the existing set of caCORE EVS services. If desired, local instances of vocabularies can be installed.
...
...
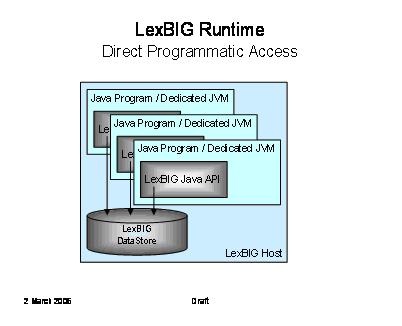
The following diagram depicts direct Java-to-Java access to LexBIG functions. This is the primary deployment scenario for phase 1.
| Info | ||
|---|---|---|
|
...
It is not required that the database be located on the same system as the program runtime. |
...
...
...
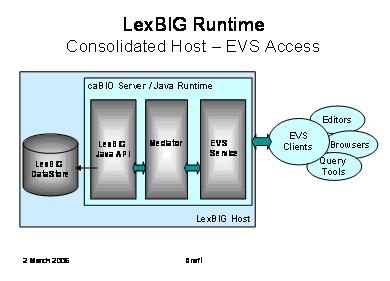
The following diagram depicts access through caCORE Enterprise Vocabulary Services (EVS) to a LexBIG vocabulary engine.
...
The primary goal is to provide a compatible experience for existing EVS browsers and client applications.
| Info | ||
|---|---|---|
|
...
This diagram shows the possible inclusion of a mediation layer between EVS and the LexBIG runtime. |
...
This would be done to facilitate alternate communications with the LexBIG server (e.g. through web services as described below). |
...
...
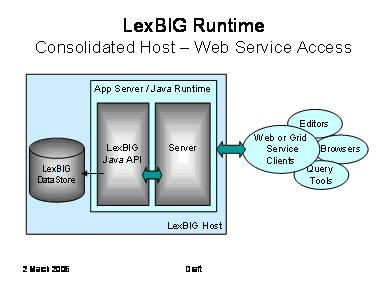
The LexBIG API is designed with web and grid-level enablement in mind. This diagram depicts deployments that wrap the current API to allow the runtime to be accessed through web or grid services.
What
...
Is LexGrid?
LexGrid is an initiative of the Mayo Clinic Division of Biomedical Informatics that focuses on the representation, storage, and dissemination of vocabularies. This effort centers on, but is not limited to, the domain of medical vocabularies and nomenclatures. Focal points of the LexGrid project include the development and promotion of standards, tools, and content that:
...
Additional information for LexGrid is available at http://informatics.mayo.edu![]() .
.
What is LexBIG?
LexBIG is a more specific project that applies LexGrid vision and technologies to requirements of the caBIG® caBIG community. The goal of the project is to build a vocabulary server accessed through a well-structured application programming interface (API) capable of accessing and distributing vocabularies as commodity resources. The server is to be built using standards-based and commodity technologies. Primary objectives for the project include:
- Provide a robust and scalable open source implementation of EVS-compliant vocabulary services. The API specification will be based on but not limited to fulfillment of the caCORE EVS API. The specification will be further refined to accommodate changes and requirements based on prioritized needs of the caBIG® caBIG community.
- Provide a flexible implementation for vocabulary storage and persistence, allowing for alternative mechanisms without impacting client applications or end users. Initial development will focus on delivery of open source freely available solutions, though this does not preclude the ability to introduce commercial solutions (e.g. Oracle).
- Provide standard tooling for load and distribution of vocabulary content. This includes but is not limited to support of standardized representations such as UMLS Rich Release Format (RRF), the OWL web ontology language, and Open Biomedical Ontologies (OBO) .
The goal for the initial year of development was to achieve the Bronze level of compatibility with regard to the caBIG® caBIG requirements. Silver-level compatibility is being pursued.</html>
| Scrollbar | ||
|---|---|---|
|