|
Page History
...
Step | Action |
|---|---|
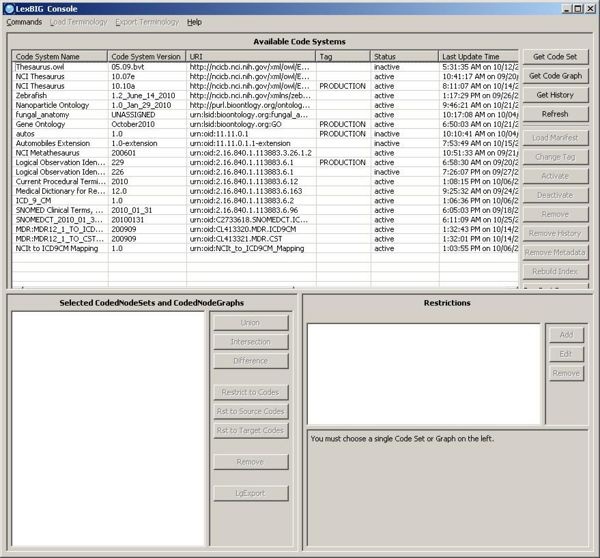
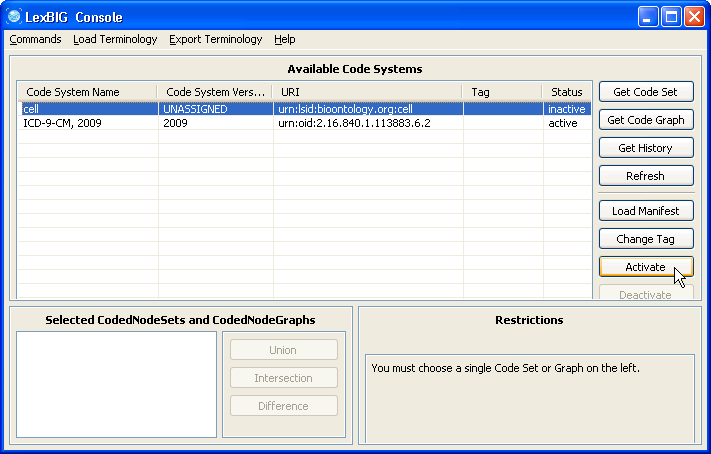
1. Launch the GUI by executing the appropriate executable for your platform. You will be presented with an application that looks like this. If the Available Code System table is empty, you will need to load at least one ontology. |
|
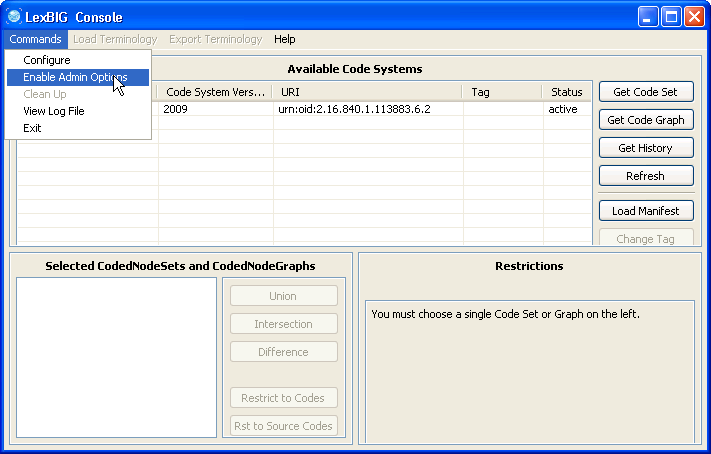
2. Selecting 'Enable Admin Options' provides administrative access to all commands. |
|
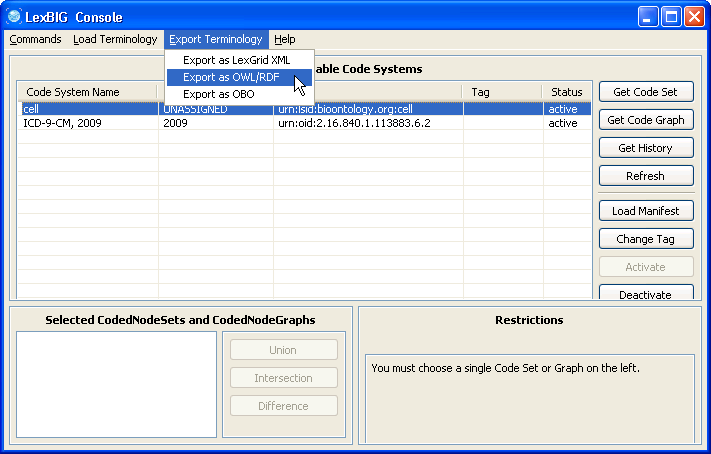
3. Select an active terminology (if the code system status is 'inactive', you can use the Activate button to activate it). Click on 'Export as OWL/RDF' button on the 'Export Terminology' menu, the exporter window will pop up. |
|
Running an OWL/RDF Export Script from the Command Line
...
Step | Action |
|---|---|
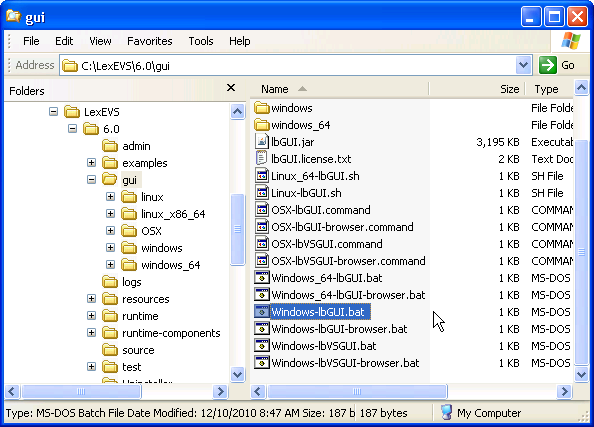
1. Launch the administration GUI. |
|
2. Enable the administration option. |
|
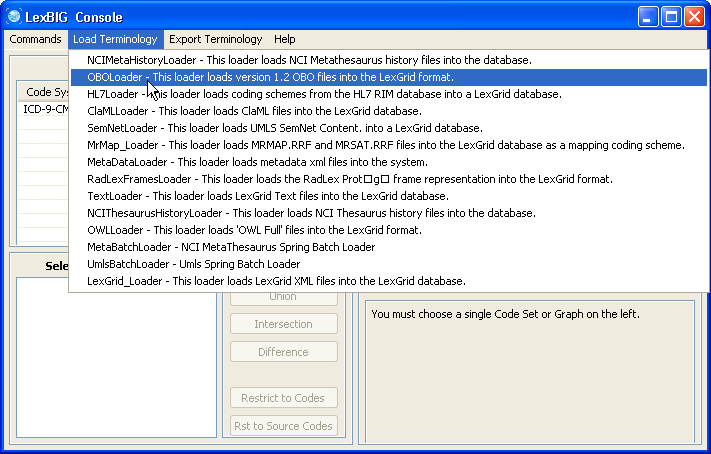
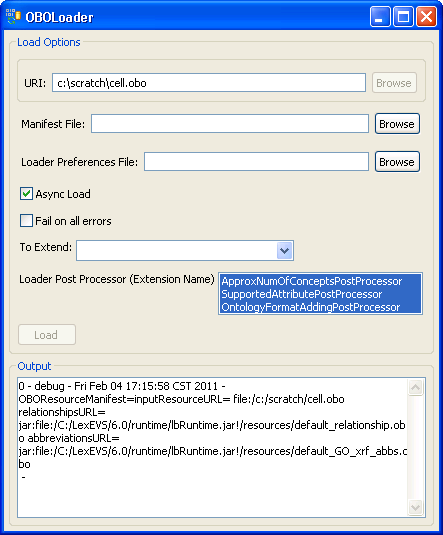
3. In this example, we use the cell.obo ontology as the example ontology, so we choose OBOloader. |
|
4. Provide cell.obo file's path. If you don't have a manifest file or preferences file then leave these fields blank. Click on the 'Load' button. After a few minutes, the loading process is done. You can find 'End State: complete' in the Output message box. Close this window, and return to the administration GUI, a 'cell' record should show up on the Available Code Systems list. |
|
5. Select the cell ontology row and click the 'Activate' button. The status column of 'cell' should change to active. |
|
6. Make sure the cell ontology is still selected. Go to the 'Export Terminology' menu, click on 'Export as OWL/RDF' in the drop down menu. |
|
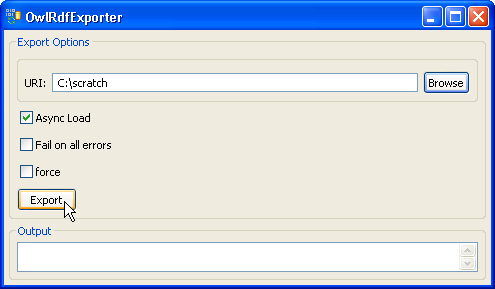
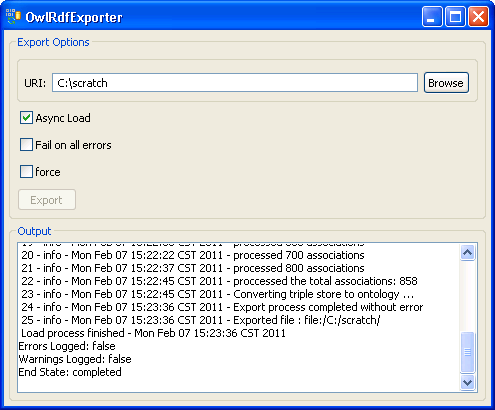
7. Provide a path in the 'URI' text box. The exported file name is automatically generated by the system, so we don't need to specify the file name. Click on the 'Export' button. |
|
8. A set of status messages will show up in the Output text box. When the 'End State: completed' statement pops up, the exporting process is complete. |
|
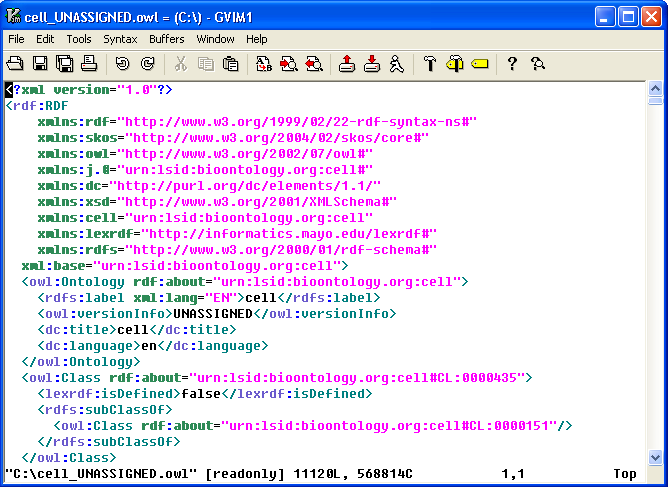
9. The convention of the exported file name is: <coding system name>_<version>.owl. In this example, cell ontology has no version information, so 'UNASSIGNED' is used as the version. The name of the OWL file in this case will be cell_UNASSIGNED.owl and it is in the C drive's root directory. |
|
...