Introduction
This document is a section of the Administration Guide.
Vocabulary Administration Overview
A set of administrative utilities are provided to manage the LexEVS Service. These utilities are provided for Windows (.bat) and Linux (.sh) operating systems. Each of the commands is located in the {LEXBIG_DIRECTORY}/admin and {LEXBIG_DIRECTORY}/test directory. A full description of the options with example is provided for each of the administration utilities.
Administrative Program |
Description |
|---|---|
ActivateScheme |
Activates a coding scheme based on unique URN and version.
|
ClearOrphanedResources |
Clean up orphaned resources - databases and indexes.
|
DeactivateScheme |
Deactivates a coding scheme based on unique URN and version.
|
ExportLgXML |
Exports content from the repository to a file in the LexGrid canonical XML format.
|
ExportOBO |
Exports content from the repository to a file in the Open Biomedical Ontologies (OBO) format.
|
ExportOWL |
Exports content from the repository to a file in OWL format.
|
ListExtensions |
List registered extensions to the LexEVS runtime environment.
|
ListSchemes |
List all currently registered vocabularies.
|
LoadLgXML |
Loads a vocabulary file, provided in LexGrid canonical xml format.
|
LoadNCIHistory |
Imports NCI History data to the LexEVS repository.
|
LoadNCIMeta |
Loads the NCI MetaThesaurus, provided as a collection of RRF files.
|
LoadNCIThesOWL |
Loads an OWL file containing a version of the NCI Thesaurus ...
|
LoadOBO |
Loads a file specified in the Open Biomedical Ontologies (OBO) format.
|
LoadOWL |
Loads an OWL file. Note Load of the NCI Thesaurus should be performed via the LoadNCIThesOWL counterpart, since it will allow more precise handling of NCI semantics. Options:
|
LoadUMLSDatabase |
Loads UMLS content, provided as a collection of RRF files in a single directory. Files may comprise the entire UMLS distribution or pruned via the MetamorphoSys tool. A complete list of source vocabularies is available online at http://www.nlm.nih.gov/research/umls/metaa1.html.
|
LoadUMLSFiles |
Loads UMLS content, provided as a collection of RRF files in a single directory. Files may comprise the entire UMLS distribution or pruned via the MetamorphoSys tool. A complete list of source vocabularies is available online at http://www.nlm.nih.gov/research/umls/metaa1.html.
|
LoadUMLSSemnet |
Loads the UMLS Semantic Network, provided as a collection of files in a single directory. The following files are expected to be provided from the National Library of Medicine (NLM) distribution:
|
LoadFMA |
Imports from an FMA database to a LexEVS repository. Requires that the pprj file be configured with a database URN, username, password for an FMA MySQL based database. The FMA.pprj file and MySQL dump file are available at http://sig.biostr.washington.edu/projects/fm/ upon registration.
|
LoadHL7RIM |
Converts an HL7 RIM MS Access database to a LexGrid database
|
LoadMetaData |
Loads optional XML-based metadata to be associated with an existing coding scheme.
|
RebuildIndex |
Rebuilds indexes associated with the specified coding scheme.
|
RemoveIndex |
Clears an optional named index associated with the specified coding scheme. Note Built-in indices required by the LexEVS runtime cannot be removed. Options:
|
RemoveScheme |
Removes a coding scheme based on unique URN and version.
|
TagScheme |
Associates a tag ID (e.g. 'PRODUCTION' or 'TEST') with a coding scheme URN and version.
|
TestRunner |
Located in {LEXBIG_DIRECTORY}/test. Executes a suite of tests for the LexEVS installation. Note The LexEVS runtime and database environments must still be configured prior to invoking the test suite. Options:
|
TransferScheme |
Tool to help gather information necessary to transfer data from one SQL server to another.
|
Installing Sample Vocabularies
This LexEVS installation provides the UMLS Semantic Net and a sampling of the NCI Thesaurus content (sample.owl) that can be loaded into the database.
- In a Command Prompt window, enter the following to go to the example programs:
cd {LEXBIG_DIRECTORY}/examples - To load the example vocabularies, run the appropriate LoadSampleData script (LoadSampleData.bat for Windows; LoadSampleData.sh for Linux).
Note
Vocabularies should not be loaded until configuration of the LexEVS runtime and database server are complete.
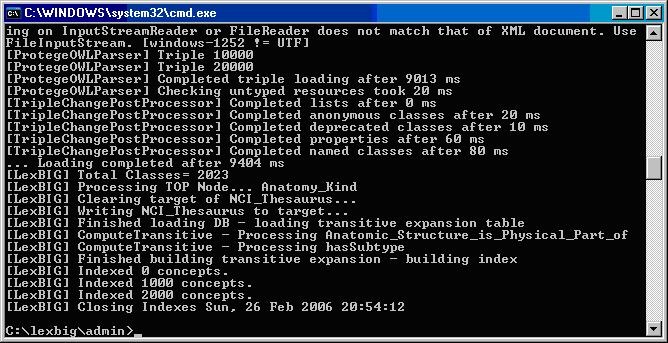
The following figure displays the successful load of the sample vocabulary file.
Running the Sample Query Programs
A set of sample programs are provided in the {LEXBIG_DIRECTORY}/examples directory. To run the sample query programs successfully a vocabulary must have been loaded.
- Enter:
cd {LEXBIG_DIRECTORY}/examples - Execute one of sample programs. .bat for windows or .sh for Linux.
- FindConceptNameForCode.bat
- FindPropsandAssocForCode.bat
- FindRelatedCodes
- FindTreeforCodeAndAssoc
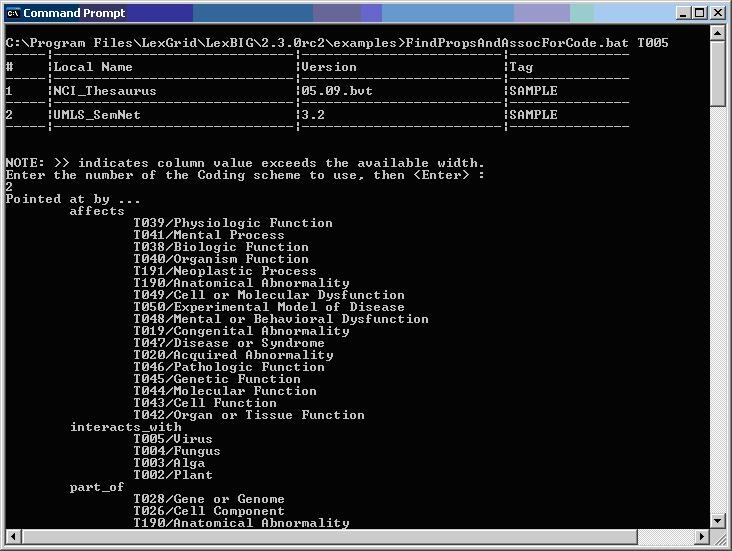
The following figure shows sample program output for finding properties and associations for a given code.
Installing NCI Vocabularies
NCI Thesaurus Vocabulary
This section describes the steps to download and install a full version of the NCI Thesaurus for the LexEVS Service.
- Using a web or ftp client go to URL: ftp://ftp1.nci.nih.gov/pub/cacore/EVS/
- Select the version of NCI Thesaurus OWL you wish to download. Save the file to a directory on your machine.
- Extract the OWL file from the zip download and save in a directory on your machine. This directory will be referred to as NCI_THESAURUS_DIRECTORY
- Using the LexEVS utilities load the NCI Thesaurus
For Windows installation use the following command:
cd {LexBIG_DIRECTORY}/adminFor Linux installation use the following command:LoadNCIThesOWL.bat -nf -in "file:///{NCI_THESAURUS_DIRECTORY}/Thesaurus_05.12f.owl"LoadNCIThesOWL.sh -nf -in "file:///{NCI_THESAURUS_DIRECTORY}/Thesaurus_05.12f.owl"Note
This step will require about three hours on a Pentium 3.0 Ghz machine. The total time to load NCI Thesaurus will vary depending on machine, memory, and disk speed.
The following example shows output from load of NCI Thesaurus 05.12f:
... [LexBIG] Processing TOP Node... Retired_Kind [LexBIG] Clearing target of NCI_Thesaurus... [LexBIG] Writing NCI_Thesaurus to target... [LexBIG] Finished loading DB - loading transitive expansion table [LexBIG] ComputeTransitive - Processing Anatomic_Structure_Has_Location [LexBIG] ComputeTransitive - Processing Anatomic_Structure_is_Physical_Part_of [LexBIG] ComputeTransitive - Processing Biological_Process_Has_Initiator_Process [LexBIG] ComputeTransitive - Processing Biological_Process_Has_Result_Biological_Process [LexBIG] ComputeTransitive - Processing Biological_Process_Is_Part_of_Process [LexBIG] ComputeTransitive - Processing Conceptual_Part_Of [LexBIG] ComputeTransitive - Processing Disease_Excludes_Finding [LexBIG] ComputeTransitive - Processing Disease_Has_Associated_Disease [LexBIG] ComputeTransitive - Processing Disease_Has_Finding [LexBIG] ComputeTransitive - Processing Disease_May_Have_Associated_Disease [LexBIG] ComputeTransitive - Processing Disease_May_Have_Finding [LexBIG] ComputeTransitive - Processing Gene_Product_Has_Biochemical_Function [LexBIG] ComputeTransitive - Processing Gene_Product_Has_Chemical_Classification [LexBIG] ComputeTransitive - Processing Gene_Product_is_Physical_Part_of [LexBIG] ComputeTransitive - Processing hasSubtype [LexBIG] Finished building transitive expansion - building index [LexBIG] Getting a results from sql (a page if using mysql) [LexBIG] Indexed 0 concepts. [LexBIG] Indexed 5000 concepts. [LexBIG] Indexed 10000 concepts. [LexBIG] Indexed 15000 concepts. [LexBIG] Indexed 20000 concepts. [LexBIG] Indexed 25000 concepts. [LexBIG] Indexed 30000 concepts. [LexBIG] Indexed 35000 concepts. [LexBIG] Indexed 40000 concepts. [LexBIG] Indexed 45000 concepts. [LexBIG] Indexed 46000 concepts. [LexBIG] Getting a results from sql (a page if using mysql) [LexBIG] Closing Indexes Mon, 27 Feb 2006 01:44:22 [LexBIG] Finished indexing
NCI Metathesaurus Vocabulary
This section describes the steps to download and install a full version of the NCI Metathesaurus for the LexEVS Service.
- Using a web or ftp client go to URL: ftp://ftp1.nci.nih.gov/pub/cacore/EVS/
- Select the version of NCI Metathesaurus RRF you wish to download. Save the file to a directory on your machine.
- Extract the RRF files from the zip download and save in a directory on your machine. This directory will be referred to as NCI_METATHESAURUS_DIRECTORY.
Note
RELEASE_INFO.RRF is required to be present for the load utility to work.
- Using the LexEVS utilities load the NCI Thesaurus
For Windows installation use the following command:
cd {LexBIG_DIRECTORY}/adminFor Linux installation use the following command:LoadNCIMeta.bat -nf -in "file:///{NCI_METATHESAURUS_DIRECTORY}/"LoadNCIMeta.sh -nf -in "file:///{NCI_THESAURUS_DIRECTORY}/"
Note
NCI Metathesaurus contains many individual vocabularies and requires several hours to load and index. This step requires about 15 hours on a Pentium 3.0 Ghz machine with 7200rpm disk. The total time to load NCI MetaThesaurus will vary depending on machine, memory, and disk speed.
NCI History
This section describes the steps to download and install a history file for NCI Thesaurus.
- Using a web or ftp client go to URL: ftp://ftp1.nci.nih.gov/pub/cacore/EVS/
- Select the version of NCI History you wish to download. Save the file to a directory on your machine. Select the VersionFile download to the same directory as the history file.
- Extract the History files from the zip download and save in a directory on your machine. This directory will be referred to as NCI_HISTORY_DIRECTORY
- Using the LexEVS utilities load the NCI Thesaurus:
For Windows installation use the following command:
cd {LexBIG_DIRECTORY}/admin_*</tt>For Linux installation use the following command:LoadNCIHistory.bat -nf -in "file:///{NCI_HISTORY_DIRECTORY}" -vf "file:///NCI_HISTORY_DIRECTORY}/VersionFile"LoadNCIHistory.sh -nf -in "file:///{NCI_HISTORY_DIRECTORY}" -vf "file:///NCI_HISTORY_DIRECTORY}/VersionFile"
Note
If a 'releaseId' occurs twice in the file, the last occurrence will be stored. If LexEVS already knows about a releaseId (from a previous history load), the information is updated to match what is provided in the file.
This file has to be provided to the load API on every load because you will need to maintain it in the future as each new release is made. We have created this file that should be valid as of today from the information that we found in the archive folder on your ftp server. You can find this file in the 'resources' directory of the LexEVS install.
Deactivating and Removing a Vocabulary
This section describes the steps to deactivate a coding scheme and remove coding scheme from LexEVS Service.
- Change directory to LexEVS administration directory. Enter:
cd {LEXBIG_DIRECTORY}/admin_* - Use the DeactiveScheme utility to prevent access to coding scheme. Once a coding scheme is deactivated, client programs will not be able to access the content for the specific coding scheme and version. Example:
DeactivateScheme -u "urn:oid:2.16.840.1.113883.3.26.1.1" -v "05.12f"
- Use RemoveScheme utility to remove coding scheme from LexEVS service and database. Example:
RemoveScheme -u "urn:oid:2.16.840.1.113883.3.26.1.1" -v "05.12f"
Tagging a Vocabulary
This section describes the steps to tag a coding scheme to be used via LexEVS API.
- Change directory to LexEVS administration directory and enter:
cd {LEXBIG_DIRECTORY}/admin_* - Use the TagScheme to tag a coding system and version with a local tag name (e.g. PRODUCTION). This tag name can be used via LexEVS API for query restriction. Example:
TagScheme -u "urn:oid:2.16.840.1.113883.3.26.1.1" -v "05.12f" -t "PRODUCTION"