
This chapter describes how to use caIntegrator tools to analyze data in subject annotation or genomic studies that have been deployed in caIntegrator.
Topics in this chaptersection include the following:

Data Analysis Overview
Once a study has been deployed, you can analyze the data using caIntegrator analysis tools.
You can verify that the study has "Deployed" status by selecting the study name in the My Studies dropdown selector. After selecting the study name, click Home in the left sidebar of the caIntegrator menu. A study summary should appear, including a status field. If the status is not deployed, or if the study summary does not appear, then the study is not deployed nor available for analysis.
If the study is ready for analysis, you will see an Analysis Tools menu in the left sidebar with the following options:
- K-M Plot: This tool analyzes subject annotation data, generating a Kaplan-Meier (K-M) plot based on survival data sets. See .
- Gene Expression Plot: This tool analyzes annotation, subject annotation or genomic data based on gene expression values. See .
- GenePattern: This feature provides an express link to GenePattern where you can perform analyses on selected caIntegrator studies, or it enables you to perform several GenePattern analyses on the grid. See .
After defining or running the analysis on selected data sets, analysis results display on the same page, allowing you to review the analysis method parameters you defined.
Creating Kaplan-Meier Plots
This topic opens from any of the three K-M plot tabs. For specific details about working with these tabs, see the following topics:
The Kaplan_Meier method analyzes comparative groups of patients or samples. In caIntegrator, the K-M method can compare survival statistics among comparative groups. You can configure the survival data in the application. For example, you might identify a group of patients with smoking history and compare survival rates with a group of non-smoking patients, or compare the survival data for two groups of patients with a specific disease type, based on Karnofsky scores. You could compare groups of patients with varying gene expression levels. You can also identify data sets using the query feature in the application, saving the queries, then configuring the K-M to compare groups identified by the queries.
The key is to first identify subsets of patients or samples that meet criteria you want to establish, thus filtering the data you want to compare. Next, generate a K-M plot based on their survival probability as a function of time. Survival differences are analyzed by the log-rank test.
caIntegrator calculates the log-rank p-Valuelog-rank p-value for the data, indicating the significance of the difference in survival between any two groups of samples. The log rank p-value is calculated using the Mantel-Haenszel method. The p-values are recalculated every time a new plot is generated.
To perform a K-M plot analysis, survival data must have been identified for the study you want to analyze or an Annotation Field Descriptor such as DAYSTODEATH has been set to Data Type 'numeric'. For more information, see on page 29. |
K-M Plot for Annotations
The groups identified for this K-M plot generation are based on annotations.
- Select the study whose data you want to analyze in the upper right portion of the caIntegrator page.
- Under Analysis Tools on the left sidebar, select K-M Plot.
- Select the For Annotation tab at the top of the page ().
- The groups to be compared in the K-M plot originate from one patient group. Varying data sets are based upon multiple values corresponding to the selected annotation. Define Patient Groups using these options:
- Annotation Type – Select the annotation type that identifies the patient group. Selections are based on the data in the chosen study.
- Annotation – Select an annotation. Fields are based on the annotation type you select. For example, if you choose Subject, then you could select Gender or Radiation Type or any field that would distinguish the patients into groups based upon their values.
- Only annotations that are defined with permissible values display in the dropdown list.
- Values – Using conventional selection techniques, select two or more values which will be the basis for the K-M plot. Permissible (available) values or "No Values" correspond to the selected annotation.
- Survival value is the length of time the patient lived. caIntegrator displays valid survival values entered for this study. Select the survival measure which is the unit of measurement for the survival value to be used for the plot.
- Click the Create Plot button.
caIntegrator generates the plot which then displays below the plot criteria ().
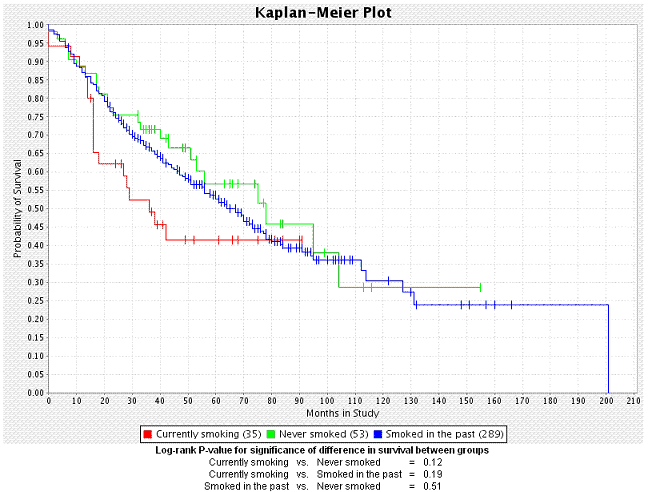
- The number of subjects for each group appears embedded in the legend of the graph below the plot.
- caIntegrator generates a P-value for the selected groups; it displays at the bottom of the page. A low P-value generally has more significance than a high P-value.
- For information regarding the P-value calculation, see .
K-M Plot for Gene Expression
caIntegrator allows you to compare expression levels for one given gene in different representative groups. The relative expression level is referred to as "fold change". Fold change is the ratio of the measured gene expression value in an experimental sample as determined by a reporter to a reference value calculated for that reporter against all control samples. The reference value is calculated by taking the mean of the log2 of the expression values for all control samples for the reporter in question. The log2 mean value  is then converted back to a comparable expression signal by returning 2 to the exponent n.
is then converted back to a comparable expression signal by returning 2 to the exponent n.
To create a K-M plot illustrating gene expression values, follow these steps:
- Select the study whose data you want to analyze in the upper right portion of the caIntegrator page. You must select a study with gene expression data.
- Under Analysis Tools on the left sidebar, select K-M Plot.
- Select the For Gene Expression tab ().

- For Gene Symbol, enter one or more gene symbols in the text box or click the icons to locate genes in the following databases. If you enter more than one gene in the text box, separate the entries by commas.
caIntegrator provides three methods whereby you can obtain gene symbols for calculating a KM plot for gene expression. For more information, see #Choosing Genes.
- Fold Change: Over-expressed/Under-expressed – Define the over- and under-expression criteria, expressed in terms of fold-change. Fold change is the ratio of the measured gene expression value for an experimental sample to the expression value for the control sample.
- Expression Level: This option allows you to run a KM gene expression plot when there is no control group nor reference data set. Enter values Above or Below Expression Level.
- Survival value – This field is required for both expression types. The length of time the patient lived. For Survival Value, select the survival measure which is the unit of measurement for the survival value to be used for the plot.
- Control Sample Sets – This field is required only for fold change data. One or more control sets are created by the study manager when a study is deployed. Select the Control Sample Set you would like to use to calculate fold-change.
If the study has more than one platform associated with it, the platform is inherently selected when you select the control set. Control sets are comprised of samples from only one platform. |
Click the Create Plot button. caIntegrator generates the plot which then displays below the plot criteria ().See .
- After you have defined the criteria as described in , caIntegrator generates the plot which then displays below the plot criteria.
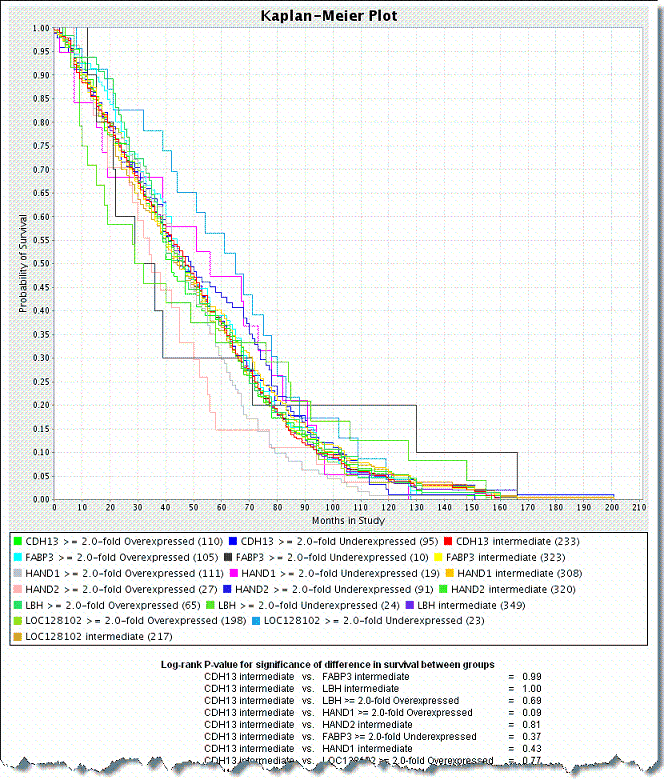
- The gene symbol for each group represented in the data appears with its color correlation to the plot embedded in the legend of the graph below the plot. Three lines on the plot represent each gene symbol entered for the plot. Each line of the three represents a subgroup of people carrying the gene--one line for overexpressed values, one line for under expressed values and one line for intermediate values which represents gene values that are not up-regulated nor down-regulated.
- In queries that include a fold change criterion and that are configured to return genomic data, raw expression values are replaced with calculated fold change values.
- A P-value is also generated for the selected groups; it displays at the bottom of the page. A low P-value generally has more significance than a high P-value.
- For information regarding the P-value calculation, see .
K-M Plot for Queries and Saved Lists
You can identify data sets using the query feature in the application. You can manipulate the queries to find the groups you want to compare, save the queries, then configure the K-M to compare the query groups. This is one method of limiting the data considered in the K-M plot calculation.
- Select the study whose data you want to analyze in the upper right portion of the caIntegrator page. The queries you identify for the K-M plot must have been saved previously in caIntegrator.
- Under Analysis Tools on the left sidebar, select K-M Plot.
- Select the For Queries and Saved Lists tab ().

- Queries – Select Queries whose data you want to analyze from the All Available Queries panel and move them to the Selected Queries panel using the Add >> button.
- Genomic queries do not appear in the lists; they cannot be selected for this type of K-M plot.
- Exclusive Subject in Queries – Check the box if you want to exclude any subjects that appear in both (or all) queries selected for the plot, thus eliminating overlap.
- Add Additional Group...all other subjects – Check the box to create an additional group of all other subjects that are not in selected query groups.
- Survival value – The length of time the patient lived. Select the survival measure which is the unit of measurement for the survival value to be used for the plot.
- Click the Create Plot button. caIntegrator generates the plot which then displays below the plot criteria ().
See .
 K-M Plot for Queries Display
K-M Plot for Queries Display
After you have defined the criteria as described in , caIntegrator generates the plot which then displays below the plot criteria.
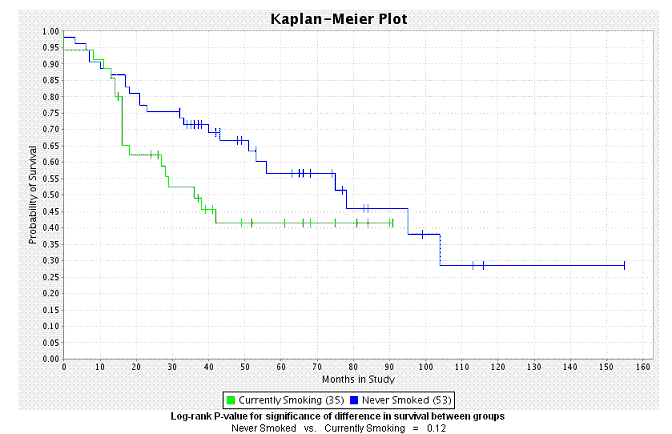
- The number of subjects for each group appears embedded in the legend of the graph below the plot.
- A P-value is also generated for the selected groups; it displays at the bottom of the page. A low P-value generally has more significance than a high P-value.
- For information regarding the P-value calculation, see .
Creating Gene Expression Plots
Gene expression plots compare signal values from reporters or genes. This statistical tool allows you to compare values for multiple genes at a time, but it does not require only two sets of data to be compared. It also allows you to compare expression levels for selected genes against expression levels for a set of control samples designated at the time of study definition.
caIntegrator provides three ways to generate meaningful gene expression plots, indicated by tabs on the page. The tabs are independent of each other and allow you to select the genes, reporters and sample groups to be analyzed on the plot.
- – You can locate genes in the caBIO directories or caIntegrator Gene Lists. You can learn more about the genes in the CGAP directory. You can define criteria for the plot using subject annotation and image annotations.
- – You can select data based on saved genomic queries.
- – You can select data based on saved subject annotation queries. You can locate genes in the caBIO directories or caIntegrator Gene Lists.
See also .
Gene Expression Value Plot for Annotation
To generate a gene expression plot, follow these steps:
- Select the study whose data you want to analyze in the upper right portion of the caIntegrator page. (You must select a study which has genomic data.)
- Under Analysis Tools on the left sidebar, select Gene Expression Plot. This opens a page with three tabs
- Select the For Annotation tab ().
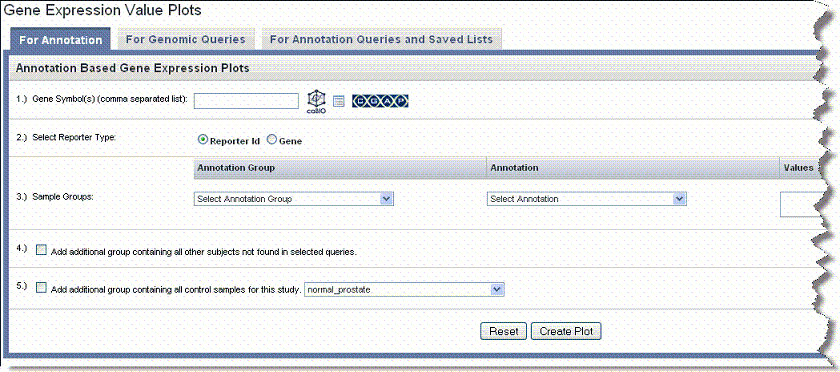
- Gene Symbol – Enter one or more gene symbols in the text box or click the icons to locate genes in the following databases. If you enter more than one gene in the text box, separate the entries by commas.
caIntegrator provides three methods whereby you can obtain gene symbols for calculating a gene expression plot. For more information, see .
- Reporter Type – Select the radio button that describes the reporter type:
- Reporter ID – Summarizes expression levels for all reporters you specify.
- Gene Name – Summarizes expression levels at the gene level.
- Platform – This field displays only if the study has multiple platforms. Select the appropriate platform for the plot. The platform you select determines the genes used for the plot.
- Sample Groups – Choose among the following options:
- Annotation Type – Select the annotation type. Selections are based on the data in the chosen study
- Annotation – Select an annotation. Fields are based on the annotation type you select. For example, if you choose Subject, then you could select Gender or Radiation Type or any field that would distinguish the patients into groups based upon study values.
- Values – Using conventional selection techniques, select one or more values which will be the basis for the plot. Permissible (available) values or "No Values" correspond to the selected annotation.
- Add Additional Group... – Define as follows:
- ...all other subjects – Check the box to create an additional group of all other subjects that are not in selected query groups.
- ...control group – Check the box to display an additional group of control samples for this study. The control set should be composed of only samples which are mapped to subjects. See on page 37.
- Click the Create Plot button. caIntegrator generates the plot which then displays below the plot criteria in bar graph format ().
See .
Gene Expression Plot for Annotation Display*
After you have defined the criteria as described in , caIntegrator generates the plot which then displays below the plot criteria.
Legends below the plot indicate the plot input. By default, the plot shows the mean of the data. displays a plot with gene expression median calculation summaries.

- You can recalculate the data display by clicking the Plot Type above the graph. See .
- You can modify the plot parameters and click the Reset button to recalculate the plot.
Gene Expression Value Plot for Genomic Queries
Data to be analyzed on this tab must have been saved as a genomic query. For more information, see on page 62.
To generate a gene expression plot using a genomic query, follow these steps:
- Select the study whose data you want to analyze in the upper right portion of the caIntegrator page. (You must select a study which has genomic data.)
- Under Analysis Tools on the left sidebar, select Gene Expression Plot.
- Select the For Genomic Queries tab ().
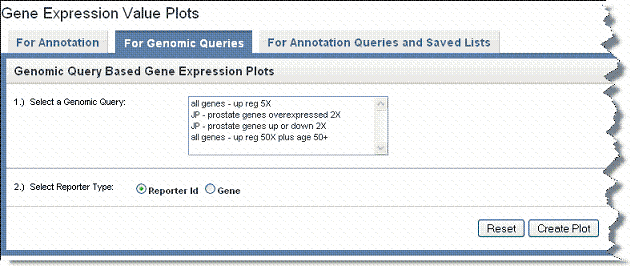
- Genomic Query – Click on the genomic query upon which the plot is to be based.
- Reporter Type – Select the radio button that describes the reporter type:
- Reporter ID – Summarizes expression levels for all reporters you specify.
- Gene Name – Summarizes expression levels at the gene level.
- Click the Create Plot button. caIntegrator generates the plot which then displays below the plot criteria. Legends below the plot indicate the plot input ().

- You can recalculate the data display by clicking the Plot Type above the graph. See .
- You can modify the plot parameters and click the Reset button to recalculate the plot.
Gene Expression Value Plot for Annotation and Saved List Queries
Data to be analyzed on this tab must have been saved as a subject annotation query, but it must have genomic data identified in the query. For more information, see on page 31. For the genomic data, you must identify genes whose expression values are used to calculate the plot.
To generate a gene expression plot using an annotation query, follow these steps:
- Select the study whose data you want to analyze in the upper right portion of the caIntegrator page. You must select a study saved as a subject annotation study, but which has genomic data.
- Under Analysis Tools on the left sidebar, select Gene Expression Plot.
- Select the For Annotation Queries and Saved Lists tab ().
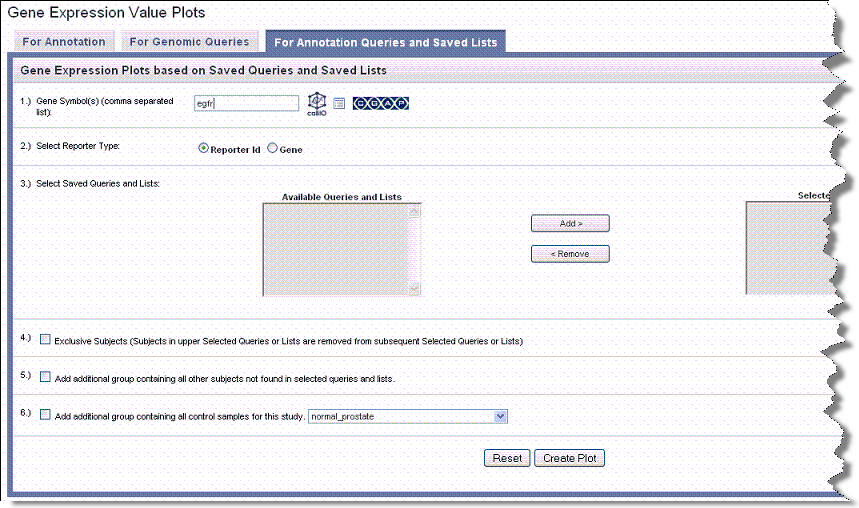
- Gene Symbol – Enter one or more gene symbols in the text box or click the icons to locate genes in the following databases. If you enter more than one gene in the text box, separate the entries by commas.
caIntegrator provides three methods whereby you can obtain gene symbols for calculating a gene expression plot. For more information, see #Choosing Genes.
- For Reporter Type, select the radio button that describes the reporter type:
- Reporter ID – Summarizes expression levels for all reporters you specify.
- Gene Name – Summarizes expression levels at the gene level.
- Platform – This field displays only if the study has multiple platforms. Select the appropriate platform for the plot. The platform you select determines the genes used for the plot.
- For Saved Queries, choose among the available saved queries and lists. Build your selections in the right panel by using the Add > and Remove < buttons.
The \[SL\] and \[Q\] prefixes to list names indicate "Subject Lists" or "Saved Queries". A "G" in the prefix indicates the list is Global. For more information, see on page 69. |
|
- Check the Exclusive Subjects... option to remove subjects in your queries and lists selection from queries or lists you use subsequently for analysis, using them exclusively for the current analysis.
- For the Add Additional Group... options, define as follows:
- ...all other subjects – Check the box to create an additional group of all other subjects that are not in selected query groups.
- ...control group – Check the box to display an additional group of control samples for this study. The control set should be composed of only samples which are mapped to subjects. See on page 37.
- Click the Create Plot button. caIntegrator generates the plot which then displays below the plot criteria in bar graph format ().
See .
Gene Expression Plot for Saved Queries Display*
After you have defined the criteria as described in , caIntegrator generates the plot which displays in bar graph format below the plot criteria.
By default, caIntegrator displays the mean of the data below the plot criteria. Legends below the plot indicate the plot input.

- You can recalculate the data display by clicking the Plot Type above the graph. See .
- You can modify the plot parameters and click the Reset button to recalculate the plot.
Understanding a Gene Expression Plot
Above the plot, you can select various plot types. When you do so, the plot is recalculated. Although all of the plots in this section appear similar, note the differences in calculation results and legends between the Y axis on each of the plots.
When you perform a Gene Expression simple search, by default the Gene Expression Plot () appears.

The Gene Expression Plot () displays mean expression intensity (Geometric mean) versus Groups.

The log2 intensity Gene Expression Plot, shown in the following figure, displays average expression intensities for the gene of interest based on Affymetrix GeneChip arrays (U133 Plus 2.0 arrays).

The box and whisker log2 expression intensity plot displays a box plot (, ). Example box and whisker plot:uses foruses of box and whisker plots include the following:
- Indicate whether a distribution is skewed and whether there are potential unusual observations (outliers) in the data set.
- Perform a large number of observations.
- Compare two or more data sets.
- Compare distributions because the center, spread, and overall range are immediately apparent.

In descriptive statistics, a box plot or boxplot, also known as a box-and-whisker diagram or plot, is a convenient way of graphically depicting groups of numerical data through their five-number summaries (the smallest observation excluding outliers, lower quartile \[Q1\], median \[Q2\], upper quartile \[Q3\], and largest observation excluding outliers). |
The box is defined by Q1 and Q3 with a line in the middle for Q2. The interquartile range, or IQR, is defined as Q3-Q1. The lines above and below the box, or 'whiskers', are at the largest and smallest non-outliers. Outliers are defined as values that are more than 1.5 * IQR greater than Q3 and less than 1.5 * IQR than Q1. Outliers, if present, are shown as open circles ().
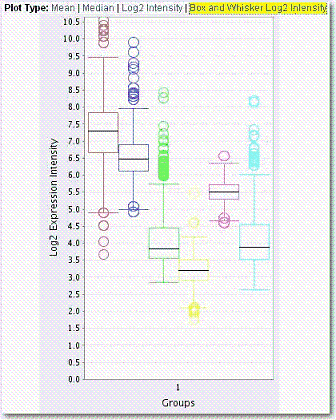
Boxplots can be useful to display differences between populations without making any assumptions of the underlying statistical distribution: they are non-parametric. The spacings between the different parts of the box help indicate the degree of dispersion (spread) and skewness in the data.

Analyzing Data with GenePattern
GenePattern is an application developed at the Broad Institute that enables researchers to access various methods to analyze genomic data. caIntegrator provides an express link to GenePattern where you can analyze data in any caIntegrator study.
Information is included in this section for connecting to GenePattern from caIntegrator. Specifics for launching GenePattern tools from caIntegrator are included as well, but you may want to refer to additional GenePattern documentation, available at this website: .
You have two options for using GenePattern from caIntegrator:
- Option 1 – Use the web-interface of any available GenePattern instances.
- To use the public instance from Broad, first register for an account at .In caIntegrator, enter the URL for connecting:[ |http://genepattern.broad.mit.edu/gp/services/], then enter your user ID and password.
- Option 2 – Use GenePattern on the grid.
The GenePattern feature in caIntegrator currently supports three analyses on the grid: Comparative Marker Selection (CMS), Principal Component Analysis (PCA) and GISTIC-supported analysis.
If you are using the web interface to access GenePattern (option #1 listed above), then you can run other GenePattern tools in addition to CMS, PCA and GISTIC. |
- Select the study whose data you want to analyze in the upper right portion of the caIntegrator page.
- Click GenePattern Analysis in the left sidebar of caIntegrator. This opens the GenePattern Analysis Status page ().
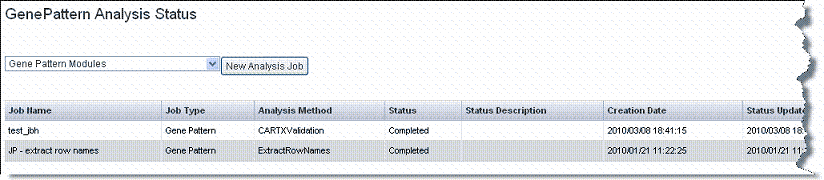
- Select from the drop-down list the type of GenePattern analysis you want to run on the data.
- GenePattern Modules – This option launches a session within GenePattern from which you can launch analyses. See .
- Comparative Marker Selection (Grid Service). This option enables you to run this GenePattern analysis on the grid. See .
- Principal Component Analysis (Grid Service). This option enables you to run this GenePattern analysis on the grid. See .
- GISTIC (Grid Service). This option enables you to run this GenePattern analysis on the grid. See .
- Click the New Analysis Job button to open a corresponding page where you can configure the analysis parameters.
GenePattern Modules
To launch the analyses described in this section, you must have a registered GenePattern account. For more information, see |
To configure the link for accessing GenePattern from caIntegrator, open the appropriate page as described in .
- Select the study whose data you want to analyze in the upper right portion of the caIntegrator page.
- Click GenePattern Analysis in the left sidebar of caIntegrator. This opens the GenePattern Analysis Status page.
- Make sure GenePattern Modules is selected in the drop down list. Click New Analysis Job.
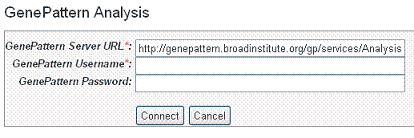
- In the GenePattern Analysis dialog box (), specify connection information, described in the following table and click Connect.
Fields |
Description |
Server URL |
Enter any GenePattern publicly available URL, such as |
GenePattern Username |
Enter your GenePattern user name. |
GenePattern Password |
Enter your GenePattern password. |
 Fields for selecting GenePattern configurations Fields for selecting GenePattern configurations
|
|
- After logging in with the GenePattern profile, the dialog box expands to includes fields for defining your GenePattern analysis.
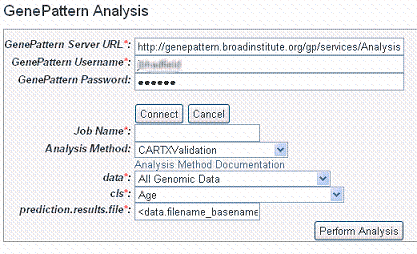
- Enter information for the following fields. Fields with a red asterisk are required:
- Job Name* – Enter a unique name for the analysis
- Analysis Method – Select any method from the drop down list. Click Analysis Method Documentation for descriptions of the different analysis methods.
- Data* – All genomic data is selected by default. Select from the list any list that has been created for this study.
- cls* – Select any annotation field
The CLS file format defines phenotype (class or template) labels and associates each sample in the expression data with a label. It uses spaces or tabs to separate the fields. The CLS file format differs somewhat depending on whether you are defining categorical or continuous phenotypes:
- Categorical labels define discrete phenotypes; for example, normal vs tumor).
- Continuous phenotypes are used for time series experiments or to define the profile of a gene of interest (gene neighbors).
- Most GenePattern modules are intended for use with categorical phenotypes. Therefore, unless the module documentation explicitly states otherwise, a CLS file should define categorical labels.
- prediction.results.file – Enter the name of this file which is part of the output from a GenePattern module.
- Click Perform Analysis. Based on the analysis method you select, you may be asked to add more information for the analysis.For more information, refer to the GenePattern Help site:
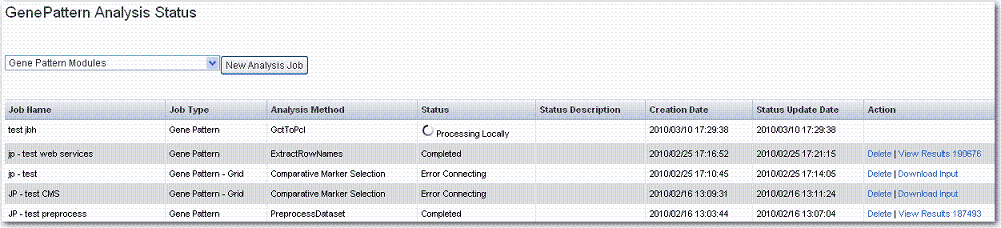
Once the analysis is launched, caIntegrator returns to the GenePattern Analysis Status page where you can monitor the status of your current study which is listed in the Analysis Method column as well as view information about other GP analyses that have been run on this study.
If you choose to access GenePattern in this way, you can continue to use GenePattern tools from within that application. See GenePattern user documentation for more information.
If you run these analyses within GenePattern itself, you may be able to view results in the GenePattern visualization module. Click View Results on the row where the results are listed. If you run them on the grid from caIntegrator, your results will be available only in spreadsheet and XML format. |
You can run GenePattern analyses for Comparative Marker Selection, Principal Component Analysis and GISTIC-based analysis on the grid if you choose.
Comparative Marker Selection (CMS) Analysis
The Comparative Marker Selection (CMS) module implements several methods to look for expression values that correlate with the differences between classes of samples. Given two classes of samples, CMS finds expression values that correlate with the difference between those two classes. If there are more than two classes, CMS can perform one-vs-all or all-pairs comparisons, depending on which option is chosen.
For more information, see the GenePattern website: .
To perform a CMS analysis, follow these steps:
- Select the study whose data you want to analyze in the upper right portion of the caIntegrator page. You must select a study saved as a subject annotation study, but which has genomic data.
- Click GenePattern Analysis in the left sidebar of caIntegrator. This opens the GenePattern Analysis Status page.
- In the GenePattern Analysis Status page, select Comparative Marker Selection (Grid Service) from the drop down list and click New Analysis Job. This opens the Comparative Marker Selection Analysis page ().

- Select or define CMS analysis parameters, described in the following table. An asterisk indicates required fields. The default settings are valid; they should provide valid results.
CMS Parameter |
Description |
Job Name* |
Assign a unique name to the analysis you are configuring. |
Preprocess Server* |
A server which hosts the grid-enabled data GenePattern PreProcess Dataset module. Select one from the list and caIntegrator will use the selected server for this portion of the processing. |
Comparative Server* |
A server which hosts the grid-enabled data GenePattern Comparative Marker Selection module. Select one from the list and caIntegrator will use the selected server for this portion of the processing. |
Annotation Queries and Lists* |
All subject annotation queries and gene lists with appropriate data for the analysis are listed. Select and move two or more queries from the All Available Queries panel to the Selected Queries panel using the Add > and Remove < buttons.
<ac:structured-macro ac:name="unmigrated-wiki-markup" ac:schema-version="1" ac:macro-id="257c70a4-3c49-49fa-9680-ccdd3803b011"><ac:plain-text-body><![CDATA[Note: The [SL] and [Q] prefixes to list names indicate "Subject Lists" or "Saved Queries". A "G" in the prefix indicates the list is Global. For more information, see on page 69. |
]]></ac:plain-text-body></ac:structured-macro> |
Filter Flag |
Variation filter and thresholding flag |
Preprocessing Flag* |
Discretization and normalization flag |
Min Change* |
Minimum fold change for filter |
Min Delta* |
Minimum delta for filter |
Threshold* |
Value for threshold |
Ceiling* |
Value for ceiling |
Max Sigma Binning* |
Maximum sigma for binning |
Probability Threshold* |
Value for uniform probability threshold filter |
Num Exclude* |
Number of experiments to exclude (max & min) before applying variation filter |
Log Base Two |
Whether to take the log base two after thresholding; default setting is "Yes". |
Number of Columns Above Threshold* |
Remove row if n columns are not >= than the given threshold
In other words, the module can remove rows in which the given number of columns does not contain a value greater or equal to a user defined threshold. |
Test Direction* |
The test to perform (up-regulated for class0; up-regulated for class1, two sided). By default, Comparative Marker Selection performs the two-sided test. |
Test Statistic* |
Select the statistic to use. |
Min Std* |
The minimum standard deviation if test statistic includes the min std option. Used only if test statistic includes the min std option. |
Number of Permutations* |
The number of permutations to perform. (Use 0 to calculate asymptotic P-values.) The number of permutations you specify depends on the number of hypotheses being tested and the significance level that you want to achieve (3). The greater the number of permutations, the more accurate the P-value.
Complete – Perform all possible permutations. By default, complete is set to No and Number of Permutations determines the number of permutations performed. If you have a small number of samples, you might want to perform all possible permutations.
Balanced – Perform balanced permutations |
Random Seed* |
The seed for the random number generator. |
Smooth P-values |
Whether to smooth P-values by using the Laplace's Rule of Succession. By default, Smooth P-values is set to Yes, which means P-values are always less than 1.0 and greater than 0.0. |
Phenotype Test* |
Tests to perform when class membership has more than 2 classes: one versus-all, all pairs.
Note: The P-values obtained from the one-versus-all comparison are not fully corrected for multiple hypothesis testing. |
- Comparative Marker Selection analysis options

|
|
- When you have completed the form, click Perform Analysis.
caIntegrator takes you to the JobStatus/Launch page where you will see the job and its status in the Status column of the list (). 
- When the job is complete, the system displays a completion date on the GenePattern Analysis status page. Click the Download link. This downloads zipped result files to your local work station. The number of files and their file type will vary according to the processing. The results format is compatible with GenePattern visualizers and can be uploaded within GenePattern.
Principal Component Analysis (PCA)
Principal Component Analysis is typically used to transform a collection of correlated variables into a smaller number of uncorrelated variables, or components. Those components are typically sorted so that the first one captures most of the underlying variability and each succeeding component captures as much of the remaining variability as possible.
You can configure GenePattern grid parameters for preprocessing the dataset in addition to PCA module parameters. For more information, see the GenePattern website: .
To perform a PCA analysis, follow these steps:
- Select the study whose data you want to analyze in the upper right portion of the caIntegrator page. You must select a study with gene expression data.
- Click GenePattern Analysis in the left sidebar of caIntegrator. This opens the GenePattern Analysis Status page.
- In the GenePattern Analysis Status page, select Principal Component Analysis (Grid Service) from the drop down list and click New Analysis Job. This opens the Principal Component Analysis page ().

- Select or define PCA analysis parameters, described in the following table. You must enter a job name and select an annotation query, but you can accept the other default settings..
PCA Parameters |
Description |
Job Name* |
Assign a unique name to the analysis you are configuring. |
Principal Component Analysis Server* |
A server which hosts the grid-enabled data GenePattern Principal Component Analysis module. Select one from the list and caIntegrator will use the selected server for this portion of the processing. |
Annotation Queries* |
All annotation queries display in this list. Select one or more of these queries to define which samples are analyzed using PCA. If you select more than one query, then the union of the samples returned by the multiple queries is analyzed. |
Cluster By* |
Selecting rows looks for principal components across all expression values, and selecting columns looks for principal components across all samples. |
- PCA analysis options

|
|
- If you want to preprocess the data set, click Enable the Preprocess Dataset. This opens an additional set of parameters (), discussed in the following table . The preprocessing is executed prior to running the PCA.

PCA Preprocessing Parameters |
Description |
Preprocess Server* |
A server which hosts the grid-enabled data GenePattern PreProcess Dataset module. Select one from the list and caIntegrator will use the selected server for this portion of the processing. |
Filter Flag |
Variation filter and thresholding flag |
Preprocessing Flag |
Discretization and normalization flag |
Min Change |
Minimum fold change for filter |
Min Delta |
Minimum delta for filter |
Threshold |
Value for threshold |
Ceiling |
Value for ceiling |
Max Sigma Binning |
Maximum sigma for binning |
Probability Threshold |
Value for uniform probability threshold filter |
Num Exclude |
Number of experiments to exclude (max & min) before applying variation filter |
Log Base Two |
Whether to take the log base two after thresholding |
Number of Columns Above Threshold |
Remove row if n columns no >= than the given threshold |
 Parameters for preprocessing data sets for PCA Parameters for preprocessing data sets for PCA
|
|
- When you have completed the form, click Perform Analysis.
- When the job is complete, the system displays a completion date on the GenePattern Analysis status page. Click the Download link. This downloads zipped result files to your local work station. The number of files and their file type will vary according to the processing. The results format is compatible with GenePattern visualizers and can be uploaded within GenePattern.
GISTIC-Supported Analysis
The GISTIC test option displays only if the study contains copy number or SNP data. For more information, see on page 38. |
The GISTIC Module is a GenePattern tool that identifies regions of the genome that are significantly amplified or deleted across a set of samples. For more information, see .
To perform a GISTIC-supported analysis, follow these steps:
- Select the study whose data you want to analyze in the upper right portion of the caIntegrator page. You must select a study with copy number (either Affymetrix SNP or Agilent Copy Number) data.
- Click GenePattern Analysis in the left sidebar of caIntegrator. This opens the GenePattern Analysis Status page.
- In the GenePattern Analysis Status page, select GISTIC (Grid Service) from the drop down list and click New Analysis Job. This opens the GISTIC Analysis page ().

- Select or define GISTIC analysis parameters, as described in the following table. You must indicate a Job Name, but you can accept the other defaults settings, which are valid and should produce valid results.
GISTIC Parameters |
Description |
Job Name* |
Assign a unique name to the analysis you are configuring. |
GISTIC Service Type* |
Select whether to use the GISTIC web service or grid service and provide or select the service address. If the web service is selected, authentication information is also required |
GenePattern User Name/Password |
Include these to log into GenePattern for the analysis. |
Annotation Queries and Lists |
All annotation queries display in this list as well as an option to select all non-control samples. Select an annotation query if you wish to run GISTIC on a subset of the data and select all non-control samples if wish to include all samples. |
Select Platform |
This option appears only if more than one copy number platform exists in the study. Select the appropriate platform from the drop-down list (). |
Exclude Sample Control Set |
From the drop-down list, select the name of the control set you want to exclude from the analysis. Click None if that is applicable. |
Amplifications Threshold* |
Threshold for copy number amplifications. Regions with a log2 ratio above this value are considered amplified. Default = 0.1. |
Deletions Threshold* |
Threshold for copy number deletions. Regions with a log2 ratio below the negative of this value are considered deletions. Default = 0.1. |
Join Segment Size* |
Smallest number of markers to allow in segments from the segmented data. Segments that contain fewer than this number of markers are joined to the neighboring segment that is closest in copy number. Default = 4. |
<ac:structured-macro ac:name="unmigrated-wiki-markup" ac:schema-version="1" ac:macro-id="dcff4b0a-65d9-436d-862d-bb3f23bcb50a"><ac:plain-text-body><![CDATA[ |
QV Thresh[hold]* |
Threshold for q-values. Regions with q-values below this number are considered significant. Default = 0.25. |
]]></ac:plain-text-body></ac:structured-macro> |
Remove X* |
Flag indicating whether to remove data from the X-chromosome before analysis. Allowed values = {1,0}. Default = 1(yes). |
cnv File |
This selection is optional.
Browse for the file. There are two options for the CNV file.
Option #1 enables you to identify CNVs by marker name. Permissible file format is described as follows:
A two column, tab-delimited file with an optional header row. The marker names given in this file must match the marker names given in the markers_file. The CNV identifiers are for user use and can be arbitrary. The column headers are:
|
- Marker Name
- CNV Identifier
Option #2 enables you to identify CNVs by genomic location. Permissible file format is described as follows:
A 6 column, tab-delimited file with an optional header row. The 'CNV Identifier', 'Narrow Region Start' and 'Narrow Region End' are for user use and can be arbitrary. The column headers are:
- CNV Identifier
- Chromosome
- Narrow Region Start
- Narrow Region End
- Wide Region Start
- Wide Region End|
- GISTIC analysis parameters
|
|
- When you have completed the form, click Perform Analysis.
- When the job is complete, the system displays a completion date on the GenePattern Analysis status page. Click the Download link. This downloads zipped result files to your local work station. The number of files and their file type will vary according to the processing. The results format is compatible with GenePattern visualizers and can be uploaded within GenePattern.
- Additionally, upon completion of a successful GISTIC anaylsis, caIntegrator automatically displays the two gene lists that it generates in the Gene List Picker so that you can use them in a caIntegrator query or plot calculation. The lists are visible only to your userID. For more information, see . The genes will also display in Saved Copy Number Analyses in the left sidebar. See on page 74.
- If samples from a copy number source are deleted, the GISTIC job in which they are appear is also deleted.
Viewing Data with the Integrative Genomics Viewer
Once you have run a query for gene expression, on page 54, or copy number data, on page 55, you can view results in the Integrative Genomics Viewer:viewing data inIntegrative Genomics Viewer (IGV).
The Integrative Genomics Viewer (IGV) is a high-performance visualization tool for interactive exploration of large, integrated datasets. It supports a wide variety of data types including sequence alignments, microarrays, and genomic annotations.
For more information about the Integrative Genomics Viewer or to connect independently to the IGV home page, click this link: .The IGV viewer and the NCI Heat Map viewer both require you to install a version of Java containing Java Web Start. For more information, see #Java for IGV and Heat Map Viewewr. |
There are two ways to integrate caIntegrator with the IGV. To configure the connection to IGV, follow one of these methods.
Method 1
- With the appropriate study open, at the bottom of the Query Results page, click the View in Integrative Genomics Viewer button.
- If you click the button at the bottom of the page with any of the query results line items selected, caIntegrator creates IGV files, with a monitor informing you of this. After the files are created, click the Launch Integrative Viewer hypertext link.
 Follow the instructions through the intermediate dialog boxes. After clicking Open with the Java program listed, the opens, displaying the dataset in the computer screen shown as follows ().
Follow the instructions through the intermediate dialog boxes. After clicking Open with the Java program listed, the opens, displaying the dataset in the computer screen shown as follows (). 
- Move your mouse to hover over the genes graphic at the bottom of the page, indicated in the figure .
- Click the mouse when you've identified a gene of interest.
This opens the genome site at UCSC , where you can learn more about the gene (). 
Go to the following website for a user guide for IGV: Method 2
- With the appropriate study open, click Integrative Genomics Viewer on the left sidebar.
- This opens the View IGV Selector page ().

- In the drop-down list, select the Gene Expression Platform for the data you want to view.
- Select the Copy Number Platform ID.
- The Annotations - Default panel displays existing annotation fields for the gene expression data in the open study. Select those fields you want to view when you open the IGV. Use the buttons for convenience if you want to Select All or Unselect All, when all are checked.
- Click View to see the data in the Integrative Genomic Viewer. caIntegrator creates IGV files of the data.
- After the files are created, click the Launch Integrative Viewer hypertext link that appears.
- Continue with .
Viewing Data with Heat Map Viewer
Heat Map Viewer:viewing data inOnce you have run a query for gene expression, on page 54, or copy number data, on page 55, you can view results in the Heat Map Viewer (HMV).
For more information about the Heat Map Viewer or to connect independently to the HMV home page, click this link: .The IGV viewer and the NCI Heat Map viewer both require you to install a version of Java containing Java Web Start. For more information, see #Java for IGV and Heat Map Viewer. |
There are two ways to integrate caIntegrator with the Heat Map Viewer. To configure the connection, follow one of these methods.
Method 1
- With the appropriate study open, at the bottom of the Query Results page, click the View in Heat Map Viewer button.
- If you click the button at the bottom of the page with any of the query results line items selected, caIntegrator creates HMV files, with a monitor informing you of this. After the files are created, click the Launch Heat Map Viewer hypertext link.
 Follow the instructions through the intermediate dialog boxes. After clicking Open with the Java program listed, the runs, displaying the dataset in the computer screen, shown as follows ().
Follow the instructions through the intermediate dialog boxes. After clicking Open with the Java program listed, the runs, displaying the dataset in the computer screen, shown as follows (). 
Go to the following website for Heat Map Viewer documentation: Method 2
- With the appropriate study open, click Heat Map Viewer on the left sidebar.
- This opens the View Heat Map Viewer Selector page ().

- Select the appropriate Copy Number Platform in the drop down list.
- The Annotations - Default panel displays existing annotation fields for the gene expression data in the open study. Select one or more annotations in the annotation list. For convenience, you can use the Select All or Unselect All buttons.
- Click View to view the data you select in Heat Map Viewer. caIntegrator creates Heat Map Viewer files of the data.
- After the files are created, click the Launch Heat Map Viewer hypertext link that appears.
- Continue with .
- For interpretation of the results and using HMV features, see the help files opened from HMV.
Java for IGV and Heat Map Viewer
To use the IGV and the NCI Heat Map viewer, described in and , you must install a version of Java containing Java Web Start. You must install recent versions of the Java Development Kit (JDK 1.5.0 aka JDK 5.0 or newer) or Java Runtime Environment (JRE 1.5.0 aka JRE 5.0 or newer). The easiest option is to install JRE 5.0. For more information, see: .
Without Java Web Start, when you click Launch Integrative Genomics Viewer or Launch Heat Map Viewer, a dialog box displays in your browser giving you the option to save or open with (IGV) or (HMV). Clicking the Open option starts the Java Web Start Launcher (default) installing the Java app so that you can view the files.
The first time you launch the IGV or HMV with Java properly installed, regardless of browser type, a warning may appear :the "the digital signature cannot be verified". Click Run to proceed with opening the viewer. |
is then converted back to a comparable expression signal by returning 2 to the exponent n.
K-M Plot for Queries Display