Author: Craig Stancl, Harold Solbrig, Scott Bauer
Email: stancl.craig@mayo.edu, solbrig.harold@mayo.edu, bauer.scott@mayo.edu
Team: LexEVS
Contract: CBITT BOA Subcontract# 29XS223
Client: NCI CBIIT
National Institutes of Heath
US Department of Health and Human Services
Revision History
Version |
Date |
Description of Changes |
Author |
|---|---|---|---|
1.0 |
5/14/10 |
Initial Version Approved via Design Review |
Team |
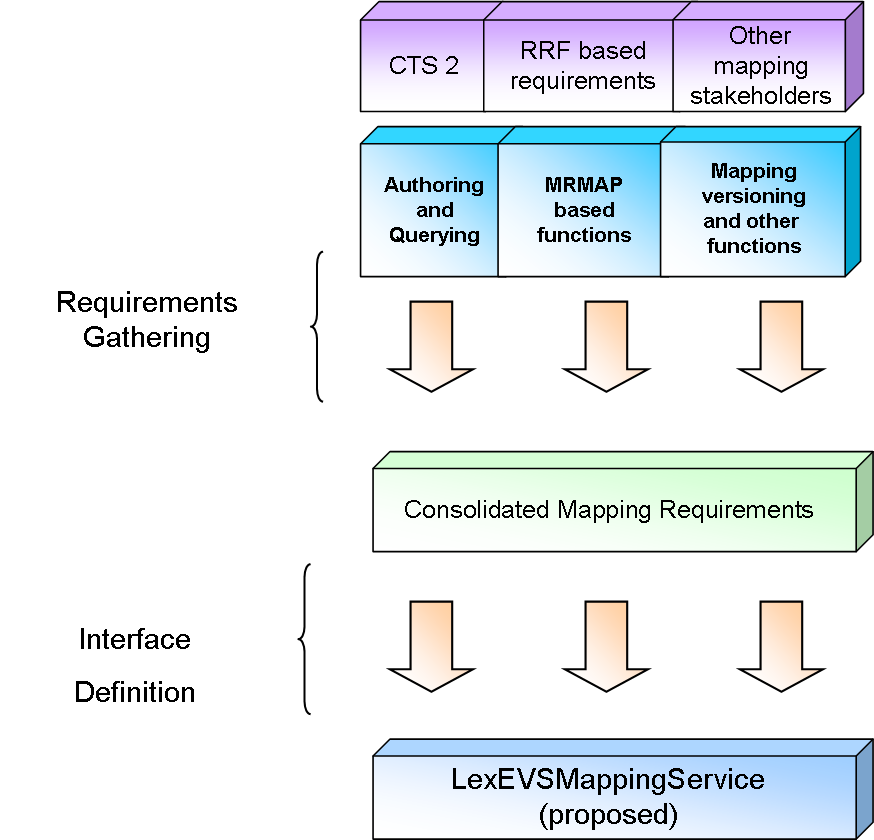
Associations and Mapping
Requirements
Levels of Mapping Implementation implied by current requirements from various stakeholders
There are multiple levels at which mapping, as evaluated here, could be considered to be "supported":
- The ability to store map sets, look up map sets by id, from, to and other attributes and to look up entries by source, target and/or relationship. We would have to store all of the additional information such as options, rules, categories, advice, etc, but it would be up to external software to interpret and evaluate the contents.
- Everything in option 1) above plus the ability to query additional fields - give me an ordered, structured list of a mapping entry in a way that someone could write a standard interpreter to evaluate it - this would basically require the addition of a model and would most likely cause us to split from the existing relations model, as it gets into the domain of table driven rule sets.
- Everything in 1) and, possibly 2) with the added ability to actually interpret the rules - an API that answers questions such as "What information is needed to map 231754000 (poisoning by sodium valproate) correctly?" (answer - whether barbituates, sulfonamides were present, whether combined with various substances, etc). "What does 231754000 in the presence of ... map to?
Evaluation of Levels of Mapping Implementation for CTS 2
For LexEVS 6.0 (CTS 2), (3) is out of scope. This is the function of a rules engine and the complexities of even the supplied examples are overwhelming. An examination of the DSS RFP may be in order to see whether there is anything there that applies. It appears that (2) is out of scope as well, as, whether the semantics of the rules are interpreted or not, the structure required to represent the rules still lies well within the rule base and DSS space. The question is, then, whether item (1) adds sufficient value to be worth doing. Our proposal is that it is not.
Since trying to fit all of the various aspects of the mapping rules that have been seen to date into the nooks and crannies of the LexGrid mapping model seems to be an exercise with little return. What is needed is to return to the set of use cases, if any, that LexEVS could reasonably resolve. As there are simpler use cases, such as those presented by various stakeholders, these use cases will need to be resolved, and if it would be useful, answer the same questions for complex mappings. Once consideration of these issues gets to the point of determining what participates in a particular map the implementation appears to be getting out of our scope - the best that might be done is supply the associationId that could be used as an index into another table.
Implications for CTS 2 Mapping Implementation
CTS 2 Function |
Description |
Scope |
|---|---|---|
Update Association Status |
Update the status of a association (active, inactive, canceled etc). This allows the ability to activate or inactivate a given association, thus changing its availability for access by other terminology service functions |
In scope for authoring API |
Create Association |
Relates a single specific coded concept within a specified code system (source) to a corresponding single specific coded concept (target) within the same or another code system, including identification of a specified Association type. |
In scope for authoring. Querying already supported |
Create Lexical Association between Coded Concepts |
Relates a set of one or more coded concepts within a specified code system (source)to a corresponding set of one or more coded concepts (target) within that system or another code system using a set of lexical rules (matching algorithms) to generate the Association. The "Source Search Criteria" allows for identification of a subset of the Source Code System to apply the matching algorithm to, if required (this may include limiting the version of the code system). |
Not in scope for CTS 2 |
Create Rules Based Association between Coded Concepts |
Relates a set of zero or more coded concepts within a specified code system (source)to a corresponding set of zero or more coded concepts (target) within that system or another code system using a set of description logic or inference rules that either assert or infer Associations. The "Source Search Criteria" allows for identification of a subset of the Source Code System to apply the matching algorithm too, if required (this may include limiting the version of the code system). |
Not in scope for CTS 2 |
Proposed Solution - Model Changes
The relations attribute in the model will have the following elements and attributes added:
- representsVersion - if present, the source asserted version of the collection of relations or mappings
- isMapping - if true, this collection of relations represents a mapping and will be evaluated for "mapping" related queries.
- sourceCodingScheme - if present, the local identifier of the namespace that the sourcEntityCodes are derived from.
- sourceCodingSchemeVersion - the source asserted version identifier of the source coding scheme. If present, this becomes the default for sourceEntityCodeNamespace.
- targetCodingScheme - if present, the local identifier of the namespace that the targetEntityCodes are derived from. If present, this becomes the default for targetEntityCodeNamespace.
- targetCodingSchemeVersion - the source asserted version identifier of the target coding scheme
- properties - all other properties on the "relations bucket" level that don't fit one of the items in the Relations container
There are no additional changes for the second part of the model, shown above.
Content Representation
MRSAT Mappings
MRSAT Column |
Function |
LexGrid Equivalent |
|---|---|---|
MAPSETVERSION |
|
Relations.representsVersion |
FROMVSAB |
The from code system version |
Relations.fromCodingSchemeVersion |
FROMRSAB |
The from code system |
Relations.FromCodingScheme |
TOVSAB |
The to code system version |
Relations.toCodingSchemeVersion |
TORSAB |
The to code system |
Relations.toCodingScheme |
MAPSETRSAB |
Source of the value set |
Relations.owner |
(anything else) |
|
Relations.property[EVS:key] |
MRMAP Mappings
There are at least two possibilities for the MRMAP transformations into LexGrid. The first, the "flattened transformation" puts the entire MRMAP row into the table.
MRMAP Flattened Option
MRMAP Column |
Function |
LexGrid Equivalent |
|---|---|---|
MAPSETCUI |
|
Relations.containerName |
MAPID |
Row identifier |
Target.associationInstanceId |
FROMEXPR |
Source code or expression |
Source.sourceEntityCode |
REL |
UMLS asserted relationship |
associationPredicate (if RELA absent) |
RELA |
Source asserted relationship |
associationPredicate(if not blank) |
TOEXPR |
Target code or expression |
Target.targetEntityCode (if code) |
TOTYPE |
Type of the data |
targetData.associationDataText.dataType (if target is not code) |
(entire MRMAP row |
|
Target(Data).associationQualification[EVS:MRMAP entry] |
MRMAP Expanded Option
MRMAP Column |
Function |
LexGrid Equivalent |
|---|---|---|
MAPSETCUI |
|
Relations.containerName |
MAPID |
Row identifier |
Target.associationInstanceId |
FROMEXPR |
Source code or expression |
Source.sourceEntityCode |
REL |
UMLS asserted relationship |
associationPredicate (if RELA absent) |
RELA |
Source asserted relationship |
associationPredicate(if not blank) |
TOEXPR |
Target code or expression |
Target.targetEntityCode (if code) |
TOTYPE |
Type of the data |
targetData.associationDataText.dataType (if target is not code) |
(anything else) |
|
Target(Data).associationQualification[EVS:associationQualifier] |
Expression Issues
One of the requirements for mapping has been the ability to discover what maps a given entity (may) participate in. This will not be completely answerable in situations where the codes are carried in text expressions. One alternativ would be to parse the target expression when possible and to create multiple rows, one for each target. Besides the parsing complexity, however, the other issue is that it is not possible to record the expression operators (AND, OR, ...) in the expansion. This, however, would arguably be the best use of the mappings for this level of complexity.
Sample Mapping XML
<?xml version="1.0" encoding="UTF-8" ?>
- <codingScheme xmlns:xsi="http://www.w3.org/2001/XMLSchema-instance" xsi:schemaLocation="http://LexGrid.org/schema/2010/01/LexGrid/codingSchemes file:/C:/devel/v6/lexgrid_model/lgModel/master/relations.xsd" xmlns="http://LexGrid.org/schema/2010/01/LexGrid/codingSchemes" xmlns:lgBuiltin="http://LexGrid.org/schema/2010/01/LexGrid/builtins" xmlns:lgCS="http://LexGrid.org/schema/2010/01/LexGrid/codingSchemes" xmlns:lgCon="http://LexGrid.org/schema/2010/01/LexGrid/concepts" xmlns:lgCommon="http://LexGrid.org/schema/2010/01/LexGrid/commonTypes" codingSchemeName="UMLS" codingSchemeURI="urn:oid:2.16.840.1.113883.6.2" representsVersion="http://SharedNames.org/ontology/umls/2009AB">
<mappings />
- <!-- MRSAT entry
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT115321814||MAPSETVERSION|MTH|2010_2009_08_17|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT115321815||TOVSAB|MTH|MSH2010_2009_08_17|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37361323||FROMRSAB|MTH|MTH|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37361327||FROMVSAB|MTH|MTH|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37361343||MAPSETGRAMMAR|MTH|ATX ::= expr; expr ::= disj; disj ::= conj , conj "OR" conj; conj ::= unary , unary "AND" unary; unary ::= neg , pos; neg ::= "NOT" pos; pos ::= "(" expr ")" , slash , atom; slash ::= atom , atom "/" atom; atom ::= "<" (any non ">" character) ">";|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37361355||MAPSETRSAB|MTH|MTH|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37361364||MAPSETTYPE|MTH|ATX|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37361368||MAPSETVSAB|MTH|MTH|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37408395||TORSAB|MTH|MSH|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37424062||SOS|MTH|This map set contains mappings from Metathesaurus CUIs to MSH associated expressions.|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT56870375||MTH_MAPFROMEXHAUSTIVE|MTH|N|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT56870378||MTH_MAPSETCOMPLEXITY|MTH|ONE_TO_ONE|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT56870384||MTH_MAPTOEXHAUSTIVE|MTH|N|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT65576316||MTH_MAPFROMCOMPLEXITY|MTH|SINGLE CUI|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT65576317||MTH_MAPTOCOMPLEXITY|MTH|BOOLEAN_EXPRESSION STR|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT67813929||MAPSETSID|MTH|1000|N||
C1306694||||CUI||AT116199817||MR|MTH|20090921|N||
C1306694||||CUI||AT31823722||DA|MTH|20040415|N||
C1306694||||CUI||AT31952620||ST|MTH|R|N||
-->
- <!-- OPTION 1: Everything flattened
-->
- <lgCS:relations containerName="C1306694" isMapping="true" sourceCodingScheme="MTH" sourceCodingSchemeVersion="MTH" targetCodingScheme="MSH" targetCodingSchemeVersion="MSH2010_2009_08_17" representsVersion="2010_2009_08_17" xmlns="http://LexGrid.org/schema/2010/01/LexGrid/relations">
- <!-- This should be MAPSETNAME but, curiously, it is missing from the MRSAT list. We are assuming that "SOS" is the equivalent
-->
<lgCommon:entityDescription>This map set contains mappings from Metathesaurus CUIs to MSH associated expressions.</lgCommon:entityDescription>
- <properties>
- <lgCommon:property propertyName="MAPSETGRAMMER">
<lgCommon:value>ATX ::= expr; expr ::= disj; disj ::= conj , conj "OR" conj; conj ::= unary , unary "AND" unary; unary ::= neg , pos; neg ::= "NOT" pos; pos ::= "(" expr ")" , slash , atom; slash ::= atom , atom "/" atom; atom ::= ">" (any non ">" character) ">";</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MAPSETRSAB">
<lgCommon:value>MTH</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MAPSETTYPE">
<lgCommon:value>ATX</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MAPSETVSAB">
<lgCommon:value>MTH</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MTH_MAPFROMEXHAUSTIVE">
<lgCommon:value>MTH</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MTH_MAPSETCOMPLEXITY">
<lgCommon:value>MTH</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MTH_MAPTOEXHAUSTIVE">
<lgCommon:value>MTH</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MTH_MAPFROMCOMPLEXITY">
<lgCommon:value>MTH</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MTH_MAPTOCOMPLEXITY">
<lgCommon:value>MTH</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MR">
<lgCommon:value>20090921</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="DA">
<lgCommon:value>20040415</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="ST">
<lgCommon:value>R</lgCommon:value>
</lgCommon:property>
</properties>
- <!-- MAPSETCUI|MAPSAB|MAPSUBSETID|MAPRANK|MAPID |MAPSID|FROMID |FROMSID|FROMEXPR|FROMTYPE|FROMRULE|FROMRES|REL|RELA
C1306694 |MTH | | |AT102971857| |C0264643| |C0264643|CUI | | |SY |
|TOID|TOSID|TOEXPR |TOTYPE |TORULE|TORES|MAPRULE|MAPRES|MAPTYPE|MAPATN|MAPATV
|3026| |<Hypertension, Renovascular> AND <Hypertension, Malignant>|BOOLEAN_EXPRESSION_STR| | | | |ATX | | ||
-->
- <associationPredicate associationName="SY">
- <!-- MRMAP Option 1 - Expand each non-semantic MRMAP entry
-->
- <source sourceEntityCode="C0264643">
- <targetData associationInstanceId="AT102971857">
- <associationQualification associationQualifier="FROMTYPE">
<qualifierText>CUI</qualifierText>
</associationQualification>
- <associationQualification associationQualifier="FROMID">
<qualifierText>C0264643</qualifierText>
</associationQualification>
- <associationQualification associationQualifier="TOTYPE">
<qualifierText>BOOLEAN_EXPRESSION_STR</qualifierText>
</associationQualification>
- <associationQualification associationQualifier="TOID">
<qualifierText>3026</qualifierText>
</associationQualification>
- <associationQualification associationQualifier="MAPTYPE">
<qualifierText>ATX</qualifierText>
</associationQualification>
<associationDataText dataType="BOOLEAN_EXPRESSION_STR">>Hypertension, Renovascular> AND >Hypertension, Malignant></associationDataText>
</targetData>
</source>
- <!-- MRMAP Option 2 - Keep MRMAP entry as it is
-->
- <source sourceEntityCode="C0264643">
- <targetData associationInstanceId="AT102971857">
- <associationQualification associationQualifier="MRMAP Entry">
<qualifierText>C1306694|MTH|||AT102971857||C0264643||C0264643|CUI|||SY||3026||>Hypertension, Renovascular> AND >Hypertension, Malignant>|BOOLEAN_EXPRESSION_STR|||||ATX||||</qualifierText>
</associationQualification>
<associationDataText dataType="BOOLEAN_EXPRESSION_STR">>Hypertension, Renovascular> AND >Hypertension, Malignant></associationDataText>
</targetData>
</source>
- <!-- TARGET Option 2 - Semantic interpretation of the target entry
-->
- <!-- NOTE That this loses the operators, but (assuming that the target entries were codes), would maintain the participation
-->
- <source sourceEntityCode="C0264643">
- <target targetEntityCode="Hyptertension, Renovascular" associationInstanceId="AT102971857.1">
- <associationQualification associationQualifier="MRMAP Entry">
<qualifierText>C1306694|MTH|||AT102971857||C0264643||C0264643|CUI|||SY||3026||>Hypertension, Renovascular> AND >Hypertension, Malignant>|BOOLEAN_EXPRESSION_STR|||||ATX||||</qualifierText>
</associationQualification>
</target>
- <target targetEntityCode="Hypertension, Malignant" associationInstanceId="AT102971857.2">
- <associationQualification associationQualifier="MRMAP Entry">
<qualifierText>C1306694|MTH|||AT102971857||C0264643||C0264643|CUI|||SY||3026||>Hypertension, Renovascular> AND >Hypertension, Malignant>|BOOLEAN_EXPRESSION_STR|||||ATX||||</qualifierText>
</associationQualification>
</target>
</source>
</associationPredicate>
</lgCS:relations>
- <!--
C2242749|L8609359|S10707041|A16492852|CODE|MTHU000002|AT110721471||FROMVSAB|MDR|MDR12_0|N||
C2242749|L8609359|S10707041|A16492852|CODE|MTHU000002|AT110721473||MAPSETVERSION|MDR|200903|N||
C2242749|L8609359|S10707041|A16492852|CODE|MTHU000002|AT110721475||MAPSETVSAB|MDR|MDR12_0|N||
C2242749|L8609359|S10707041|A16492852|CODE|MTHU000002|AT90562335||FROMRSAB|MDR|MDR|N||
C2242749|L8609359|S10707041|A16492852|CODE|MTHU000002|AT90562339||MAPSETRSAB|MDR|MDR|N||
C2242749|L8609359|S10707041|A16492852|CODE|MTHU000002|AT90562341||MAPSETTYPE|MDR|MedDRA to ICD9CM Mappings|N||
C2242749|L8609359|S10707041|A16492852|CODE|MTHU000002|AT90580448||MTH_MAPFROMCOMPLEXITY|MDR|SINGLE CODE|N||
C2242749|L8609359|S10707041|A16492852|CODE|MTHU000002|AT90580450||MTH_MAPFROMEXHAUSTIVE|MDR|N|N||
C2242749|L8609359|S10707041|A16492852|CODE|MTHU000002|AT90580452||MTH_MAPSETCOMPLEXITY|MDR|ONE_TO_ONE|N||
C2242749|L8609359|S10707041|A16492852|CODE|MTHU000002|AT90580454||MTH_MAPTOCOMPLEXITY|MDR|SINGLE CODE|N||
C2242749|L8609359|S10707041|A16492852|CODE|MTHU000002|AT90580456||MTH_MAPTOEXHAUSTIVE|MDR|N|N||
C2242749|L8609359|S10707041|A16492852|CODE|MTHU000002|AT91157709||TORSAB|MDR|ICD9CM|N||
C2242749|L8609359|S10707041|A16492852|CODE|MTHU000002|AT91157711||TOVSAB|MDR|ICD9CM_1998|N||
C2242749||||CUI||AT101551784||DA|MTH|20081103|N||
C2242749||||CUI||AT102850765||ST|MTH|R|N||
C2242749||||CUI||AT116202416||MR|MTH|20090926|N||
-->
- <!-- Should this be the code or "MTHU000002"?
-->
- <lgCS:relations containerName="C2242749" sourceCodingScheme="MDR" sourceCodingSchemeVersion="MDR12_0" targetCodingScheme="ICD9CM" targetCodingSchemeVersion="ICD9CM_1998" representsVersion="200903" xmlns="http://LexGrid.org/schema/2010/01/LexGrid/relations">
<lgCommon:owner>MDR</lgCommon:owner>
- <!-- This should be MAPSETNAME but, curiously, it is missing from the MRSAT list
-->
<lgCommon:entityDescription>This map set contains mappings from Metathesaurus CUIs to MSH associated expressions.</lgCommon:entityDescription>
- <properties>
- <lgCommon:property propertyName="MAPSETTYPE">
<lgCommon:value>MedDRA to ICD9CM Mappings</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MTH_MAPFROMCOMPLEXITY">
<lgCommon:value>SINGLE CODE</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MTH_MAPFROMEXHAUSTIVE">
<lgCommon:value>N</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MTH_MAPSETCOMPLEXITY">
<lgCommon:value>ONE_TO_ONE</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MTH_MAPTOCOMPLEXITY">
<lgCommon:value>SINGLE CODE</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MTH_MAPTOEXHAUSTIVE">
<lgCommon:value>MedDRA to ICD9CM Mappings</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MAPSETTYPE">
<lgCommon:value>N</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="DA">
<lgCommon:value>20081103</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="ST">
<lgCommon:value>R</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MR">
<lgCommon:value>20090926</lgCommon:value>
</lgCommon:property>
</properties>
- <!-- MAPSETCUI|MAPSAB|MAPSUBSETID|MAPRANK|MAPID |MAPSID|FROMID |FROMSID|FROMEXPR|FROMTYPE|FROMRULE|FROMRES|REL|RELA
C2242749 |MDR | | |AT91157713 | |10032725| |10032725|CODE | | |RQ |mapped_to
|TOID |TOSID|TOEXPR |TOTYPE |TORULE|TORES|MAPRULE|MAPRES|MAPTYPE|MAPATN|MAPATV
|728.7| |728.7 |CODE |||||||||
-->
- <associationPredicate associationName="mapped_to">
- <!-- Note that "RQ" gets lost in this approach...
-->
- <source sourceEntityCode="10032725">
- <target targetEntityCode="728.7" associationInstanceId="AT91157713">
- <associationQualification associationQualifier="FROMTYPE">
<qualifierText>CODE</qualifierText>
</associationQualification>
- <associationQualification associationQualifier="FROMID">
<qualifierText>10032725</qualifierText>
</associationQualification>
- <associationQualification associationQualifier="TOID">
<qualifierText>728.7</qualifierText>
</associationQualification>
</target>
</source>
</associationPredicate>
</lgCS:relations>
- <!-- C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT106958256||MAPSETRSAB|ICD10PCS|ICD10PCS|N||
C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT106958265||MTH_MAPFROMCOMPLEXITY|ICD10PCS|SINGLE SDUI|N||
C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT106958266||MTH_MAPFROMEXHAUSTIVE|ICD10PCS|N|N||
C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT106958268||MTH_MAPSETCOMPLEXITY|ICD10PCS|N_TO_N|N||
C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT106958270||MTH_MAPTOEXHAUSTIVE|ICD10PCS|N|N||
C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT106958273||TORSAB|ICD10PCS|ICD10PCS|N||
C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT112240306||FROMRSAB|ICD10PCS|ICD9CM|N||
C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT112240309||FROMVSAB|ICD10PCS|ICD9CM_2009|N||
C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT112240314||MAPSETTYPE|ICD10PCS|ICD-9-CM to ICD-10-PCS Mappings (GEMs)|N||
C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT112240316||MAPSETVERSION|ICD10PCS|2009|N||
C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT112240319||MAPSETVSAB|ICD10PCS|ICD10PCS_2009|N||
C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT112240322||MAPSETXRTARGETID|ICD10PCS|NoPCS|N||
C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT112240327||MTH_MAPTOCOMPLEXITY|ICD10PCS|SINGLE SCUI|N||
C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT112240330||SOS|ICD10PCS|This set maps ICD-9-CM codes to ICD-10-PCS. These are "General Equivalence Mappings" (GEMs) and are rule-based.|N||
C2603385|L8770856|S10872372|A16736317|CODE|MTHU000001|AT112240336||TOVSAB|ICD10PCS|ICD10PCS_2009|N||
C2603385||||CUI||AT109499260||DA|MTH|20090401|N||
C2603385||||CUI||AT110315629||ST|MTH|R|N||
C2603385||||CUI||AT116202419||MR|MTH|20090926|N||
-->
- <lgCS:relations containerName="C2603385" sourceCodingScheme="ICD9CM" sourceCodingSchemeVersion="ICD9CM_2009" targetCodingScheme="ICD10PCS" targetCodingSchemeVersion="ICD10PCS_2009" representsVersion="ICD10PCS_2009" xmlns="http://LexGrid.org/schema/2010/01/LexGrid/relations">
<lgCommon:owner>ICD10PCS</lgCommon:owner>
<lgCommon:entityDescription>This set maps ICD-9-CM codes to ICD-10-PCS. These are "General Equivalence Mappings" (GEMs) and are rule-based.</lgCommon:entityDescription>
- <properties>
- <lgCommon:property propertyName="MTH_MAPFROMCOMPLEXITY">
<lgCommon:value>SINGLE SDUI</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MTH_MAPFROMEXHAUSTIVE">
<lgCommon:value>N</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MTH_MAPSETCOMPLEXITY">
<lgCommon:value>N_TO_N</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MTH_MAPTOEXHAUSTIVE">
<lgCommon:value>N</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MAPSETVERSION">
<lgCommon:value>2009</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MAPSETXRTARGETID">
<lgCommon:value>NoPCS</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MTH_MAPTOCOMPLEXITY">
<lgCommon:value>SINGLE SCUI</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="DA">
<lgCommon:value>20090401</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="ST">
<lgCommon:value>R</lgCommon:value>
</lgCommon:property>
- <lgCommon:property propertyName="MR">
<lgCommon:value>20090926</lgCommon:value>
</lgCommon:property>
</properties>
- <!-- MAPSETCUI|MAPSAB |MAPSUBSETID|MAPRANK|MAPID |MAPSID|FROMID |FROMSID|FROMEXPR|FROMTYPE|FROMRULE|FROMRES|REL|RELA
C2603385 |ICD10PCS|0:0 | |AT106958276| |86.63 | |86.63 |SDUI | | |RO |approximately_mapped_to
|TOID |TOSID|TOEXPR |TOTYPE |TORULE|TORES|MAPRULE|MAPRES|MAPTYPE|MAPATN|MAPATV
|0HR6X73| |0HR6X73|SCUI |||||||||
-->
- <associationPredicate associationName="approximately_mapped_to">
- <!-- Note that "RO" gets lost in this approach...
-->
- <source sourceEntityCode="86.63">
- <target targetEntityCode="0HR6X73" associationInstanceId="AT106958276">
- <!-- NOTE: This does NOT imply any ordering in the way that map entries are returned !
-->
- <associationQualification associationQualifier="MAPSUBSETID">
<qualifierText>0:0</qualifierText>
</associationQualification>
- <associationQualification associationQualifier="FROMTYPE">
<qualifierText>SDUI</qualifierText>
</associationQualification>
- <associationQualification associationQualifier="FROMID">
<qualifierText>86.63</qualifierText>
</associationQualification>
- <associationQualification associationQualifier="TOTYPE">
<qualifierText>SCUI</qualifierText>
</associationQualification>
- <associationQualification associationQualifier="TOID">
<qualifierText>0HR6X73</qualifierText>
</associationQualification>
</target>
</source>
</associationPredicate>
- <associationPredicate associationName="XR">
- <!-- Note that approximately_mapped_to goes away. This may not be what we want?
-->
- <!-- MAPSETCUI|MAPSAB |MAPSUBSETID|MAPRANK|MAPID |MAPSID|FROMID |FROMSID|FROMEXPR|FROMTYPE|FROMRULE|FROMRES|REL|RELA
C2603385 |ICD10PCS|0:0 | |AT112240364| |89.03 | |89.03 |SDUI | | |XR ||||||||||||||
|TOID |TOSID|TOEXPR |TOTYPE |TORULE|TORES|MAPRULE|MAPRES|MAPTYPE|MAPATN|MAPATV
|||||||||||||
-->
- <source sourceEntityCode="89.03">
- <target targetEntityCode="NoPCS" associationInstanceId="AT112240364">
- <associationQualification associationQualifier="MAPSUBSETID">
<qualifierText>0:0</qualifierText>
</associationQualification>
- <associationQualification associationQualifier="FROMTYPE">
<qualifierText>SDUI</qualifierText>
</associationQualification>
- <associationQualification associationQualifier="FROMID">
<qualifierText>86.63</qualifierText>
</associationQualification>
- <associationQualification associationQualifier="TOID">
<qualifierText>NoPCS</qualifierText>
- <!-- This is derived from the MAPSETXRTARGETID property in MRSAT
-->
</associationQualification>
</target>
</source>
</associationPredicate>
</lgCS:relations>
</codingScheme>
A UMLS Example
Taking the first row in the MRMAP.RRF table (UMLS 2009AB) as an example, we have:
C1306694|MTH|||AT102971857||C0264643||C0264643|CUI|||SY||3026||<Hypertension, Renovascular> AND <Hypertension, Malignant>|BOOLEAN_EXPRESSION_STR|||||ATX||||
And the corresponding MRSAT rows for this mapping are:
CUI LUI SUI METAUI STYPE CODE ATUI ATN SAB ATV SUPPRESS CVF
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT115321814||MAPSETVERSION|MTH|2010_2009_08_17|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT115321815||TOVSAB|MTH|MSH2010_2009_08_17|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37361323 ||FROMRSAB|MTH|MTH|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37361327 ||FROMVSAB|MTH|MTH|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37361343 ||MAPSETGRAMMAR|MTH|ATX ::= expr; expr ::= disj; disj ::= conj , conj "OR" conj; conj ::= unary , unary "AND" unary; unary ::= neg , pos; neg ::= "NOT" pos; pos ::= "(" expr ")" , slash , atom; slash ::= atom , atom "/" atom; atom ::= "<" (any non ">" character) ">";|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37361355 ||MAPSETRSAB|MTH|MTH|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37361364 ||MAPSETTYPE|MTH|ATX|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37361368 ||MAPSETVSAB|MTH|MTH|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37408395 ||TORSAB|MTH|MSH|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT37424062 ||SOS|MTH|This map set contains mappings from Metathesaurus CUIs to MSH associated expressions.|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT56870375 ||MTH_MAPFROMEXHAUSTIVE|MTH|N|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT56870378 ||MTH_MAPSETCOMPLEXITY|MTH|ONE_TO_ONE|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT56870384 ||MTH_MAPTOEXHAUSTIVE|MTH|N|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT65576316 ||MTH_MAPFROMCOMPLEXITY|MTH|SINGLE CUI|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT65576317 ||MTH_MAPTOCOMPLEXITY|MTH|BOOLEAN_EXPRESSION STR|N||
C1306694|L8923988|S11111536|A17029750|CODE|1000|AT67813929 ||MAPSETSID|MTH|1000|N||
C1306694| | | |CUI | |AT116199817||MR|MTH|20090921|N||
C1306694| | | |CUI | |AT31823722 ||DA|MTH|20040415|N||
C1306694| | | |CUI | |AT31952620 ||ST|MTH|R|N||
The corresponding Relations container would have the following characteristics
containerName: (TBD)
entityDescription: This map set contains mappings from Metathesaurus CUIs to MSH associated expressions.
representsVersion: 2010_2009_07_17
isMapping: true
sourceCodingScheme: MTH
sourceCodingSchemeVersion: MTH (or nothing?)
targetCodingScheme: MSH
targetCodingSchemeVersion: MSH2010_2009_08_17
property: MAPSETGRAMMAR
value: ATX ::= expr; expr ::= disj; disj ::= conj , conj "OR" conj; conj ::= unary , unary "AND" unary; unary ::= neg , pos; neg ::= "NOT" pos; pos ::= "(" expr ")" , slash , atom; slash ::= atom , atom "/" atom; atom ::= "<" (any non ">" character) ">";
Note - it might be handy to use the dataType field here to encode the grammer (BNF?))
property: MAPSETRSAB
value: MTH
property: MAPSETTYPE
value: ATX
property: MAPSETVSAB
value: MSH
property: MTH_MAPFROMEXHAUSTIVE
value: N
property: MTH_MAPSETCOMPLEXITY
value: N
property: MTH_MAPTOEXHAUSTIVE
value: N
property: MTH_MAPTOCOMPLEXITY
value: BOOLEAN_EXPRESSION_STRING
property: MR
value: 20090921
property: DA
value: 20040415
property: ST
value: R
Note: it may be that the last couple of fields may map into the versionable characteristics of the relations container
value: 20090921
The MRMAP entry below is an example of a mapping that is complex enough that it can't be completely interpreted by the existing API
MAPSETCUI|MAPSETSAB| |MAPSID ||FROMEXPR||FROMRULE| RELA TOSID|TOEXPR |TOTYPE C1306694 |MTH |||AT102971857||C0264643||C0264643|CUI|||SY||3026||<Hypertension, Renovascular> AND <Hypertension, Malignant>|BOOLEAN_EXPRESSION_STR|||||ATX|||| associationPredicate.associationName: SY associationSource.sourceEntityCode: C0264643 associationTarget.targetEntityCode: Hypertension, Renovascular associationQualification.qualifierName: MAPSID associationQualification.qualifierText: AT10297857 associationQualification.qualifierName: FROMRULE associationQualification.qualifierText: C026463 associationQualification.qualifierName: FROMEXPR associationQualification.qualifierText: C0264643 associationQualification.qualifierName: FROMRULE associationQualification.qualifierText: C0264643 associationQualification.qualifierName: FROMTYPE associationQualification.qualifierText: CUI associationQualification.qualifierName: TOSID associationQualification.qualifierText: 3026 associationQualification.qualifierName: TOEXPR associationQualification.qualifierText: <Hypertension, Renovascular> AND <Hypertension, Malignant> associationQualification.qualifierName: TOTYPE associationQualification.qualifierText: BOOLEAN_EXPRESSION associationTarget.targetEntityCode: Hypertension, Malignant associationQualification.qualifierName: MAPSID associationQualification.qualifierText: AT10297857 associationQualification.qualifierName: FROMRULE associationQualification.qualifierText: C026463 associationQualification.qualifierName: FROMEXPR associationQualification.qualifierText: C0264643 associationQualification.qualifierName: FROMRULE associationQualification.qualifierText: C0264643 associationQualification.qualifierName: FROMTYPE associationQualification.qualifierText: CUI associationQualification.qualifierName: TOSID associationQualification.qualifierText: 3026 associationQualification.qualifierName: TOEXPR associationQualification.qualifierText: <Hypertension, Renovascular> AND <Hypertension, Malignant> associationQualification.qualifierName: TOTYPE associationQualification.qualifierText: BOOLEAN_EXPRESSION
An alternative representation would be:
associationTarget.targetEntityCode: Hypertension, Malignant associationQualification.qualifierName: MRMAP_ENTRY associationQualification.qualifierText: C1306694 |MTH |||AT102971857||C0264643||C0264643|CUI|||SY||3026||<Hypertension, Renovascular> AND <Hypertension, Malignant>|BOOLEAN_EXPRESSION_STR|||||ATX|||| associationTarget.targetEntityCode: Hypertension, Renovascular associationQualification.qualifierName: MRMAP_ENTRY associationQualification.qualifierText: C1306694 |MTH |||AT102971857||C0264643||C0264643|CUI|||SY||3026||<Hypertension, Renovascular> AND <Hypertension, Malignant>|BOOLEAN_EXPRESSION_STR|||||ATX||||
Summary of Mapping Implementation for LexEVS 6.0 and CTS 2
CTS 2 Function |
Description |
|---|---|
Update Association Status |
Update the status of a association (active, inactive, canceled etc). This allows the ability to activate or inactivate a given association, thus changing its availability for access by other terminology service functions |
Create Association |
Relates a single specific coded concept within a specified code system (source) to a corresponding single specific coded concept (target) within the same or another code system, including identification of a specified Association type. |
Persist MRMAP data |
Creates association and qualifier for the given concept as target establishing a "map entry" with unprocessed text string from MRMAP row as qualifier value. Other values for association predicate and source pulled from MRMAP as required. |