- Documentation Main Page
- Creation of the Cancer Gene Index
- Data, Metadata, and Annotations
- Cancer Gene Index Gene-Disease and Gene-Compound XML Documents
- caBIO APIs
- Cancer Gene Index Shared Parsed Data and Code
- caBIO Portlet Templated Searches
- caBIO Home Page
- caBIO iPhone Application
- caBIO Portlet Simple Searches
- Glossary
- Credits and Resources
Need Additional Help?
If you need support, please contact Application Support.
To Print the Guide
We recommend you print one wiki page of the guide at a time. To do this, click the printer icon at the top right of the page; then from the browser File menu, choose Print. Printing multiple pages at one time is more complex. For instructions, refer to How do I print multiple pages?.
Having Trouble Reading the Text?
Resizing the text for any web page is easy. For information on how to do this in your web browser, refer to this W3C tutorial .
Selecting a Gene Search Term
The caBIO Portlet Templated Search tool allows end users to execute case-insensitive searches for the diseases and compounds that are associated with a gene concept. These search terms must match exact gene terms in the Cancer Gene Index.
If you would like to search for a specific gene (for example, only for "BMP7"), you may use the gene's HUGO gene symbol .
If you would like to search for diseases and compounds that are associated with a related genes (for example, "BMP2," "BMP3," "BMP5", and "BMP7"), use the * wild card character, which matches zero or more characters, in your search terms (for example, "BMP*"). For example, a search for "bmp7" will return nearly 210 results that are related to that exact term, whereas a search for "bmp*" returns nearly 2,200 results, where genes match terms that include "BMP1," "BMP2," and "BMPR2."
Using Templated Search Tool to Retrieve Gene-Disease and Gene-Compound Concept Pairs
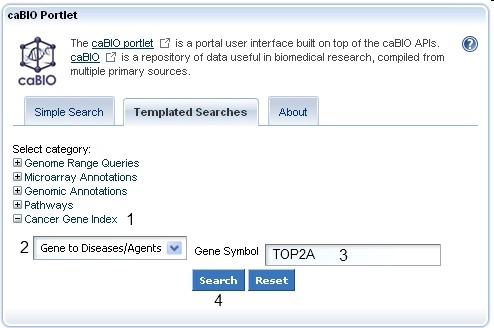
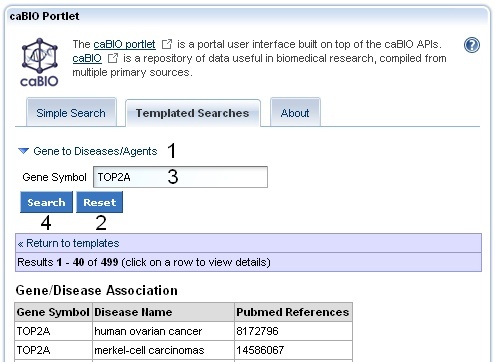
To search for Cancer Gene Index gene-disease and gene-compound data on the Templated Search page, click on the Cancer Gene Index Category node (1) and choose the Gene to Diseases/Agents query option (2). Manually delete the suggested search term "C4BPA" (3), enter your compound term or concept code into the text box (for example, "BMP7" or "BMP*"), and click the Search button (4).
Note
Although the Cancer Gene Index refers to pharmacological substances as "compounds," caBIO refers to the concept as "agents."
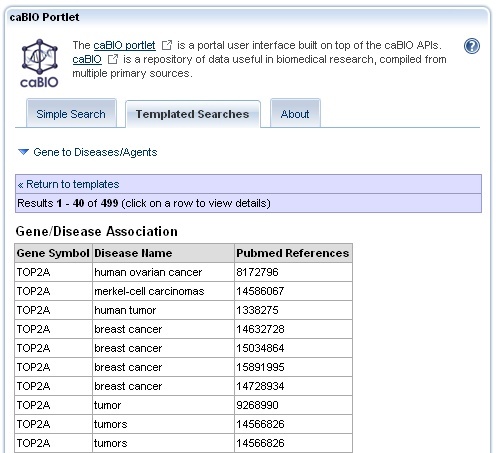
The Gene to Diseases/Agents query returns Gene/Agent Association and Gene/Disease Association results in a tabular form. Each table consists of three columns which include: HUGO Gene Symbol, NCI Thesaurus disease or compound term, and a PubMed identifier. Each row represents a single gene-disease or gene-compound association. You will will receive a "No results found" message if you have used a search term that does not match any Cancer Gene Index gene symbol or if no disease or compound is associated with your gene search term.
Warning!
Your first result page will likely contain a Gene/Agent Association and Gene/Disease Association table. This does not mean that all of the gene-compound associations are given on the first page. Instead you must continue to review subsequent results pages to find all gene-compound and gene-disease association results.
Saving Your Search Results
To save your search results, copy and paste them into a text editor or spreadsheet.
Validating Your Results
caBIO houses the all Cancer Gene Index gene-disease association annotations, including information on whether the association was found to be negative by the Cancer Gene Index project's human curators or originated from cell lines or non-human organisms. In order to select only your desired data, you must access to caBIO Object Graph Browser to manually check the negation status of and find other annotations for each retrieved record.
Validating Your Retrieved Compound Results
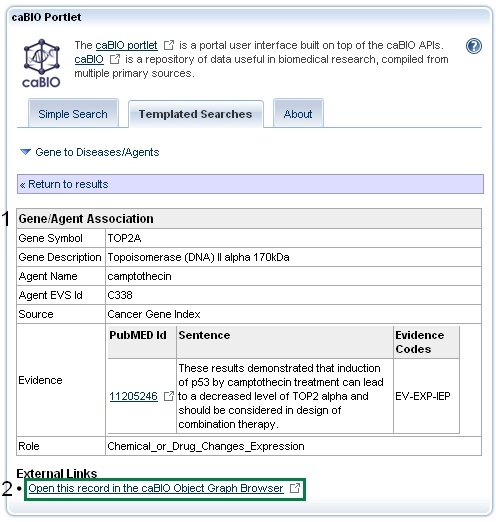
To perform this validation, click on any gene-compound result row in order to view its Gene-Agent Association information. Gene-Agent Association data include the Gene Term, Gene Symbol, Agent Term, Agent Identifier, Sentence PubMed Identifier, Sentence Evidence, Evidence Code, and Role Code or Detail (1).
Next, click on the Open this record in the caBIO Object Graph Browser at the bottom of the page (2).
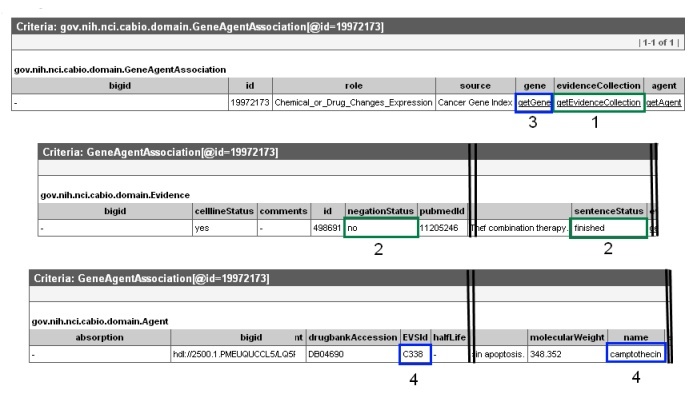
This will open up a the Gene-Agent Association object in a new window or tab in your web browser. Click on the getEvidenceCollection link (3, green box) to open the full Evidence record, which contains all of the annotations made by the curator for that particular piece of evidence of a gene-compound association. Gene-compound associations with true relationships were validated as being true associations by human curators have a sentenceStatus set to "finished" and a negationStatus set to "no" (4, middle and right green boxes). If you wish to exclude data from cell lines, for example, the celllineStatus must be be "no." If you want to see additional information about the agent, navigate back to the Gene-Agent Association in your browser and click on getAgent (3, blue box in top panel). This will open an Agent object, in which you may see the name and EVS Identifier for the associated compound (4, blue boxes in bottom panel).
Note
If you accidentally navigate to somewhere else in caBIO by clicking on the wrong link in the Object Viewer and are confused, stop and go back!
Once you have finished reviewing the gene-compound association in the caBIO Object Graph Browser, return to the caBIO Portlet web browser tab or window, and click the Return to Results link in blue to continue evaluating retrieved gene-compound pairs in the results table.
Validating Your Retrieved Disease Results
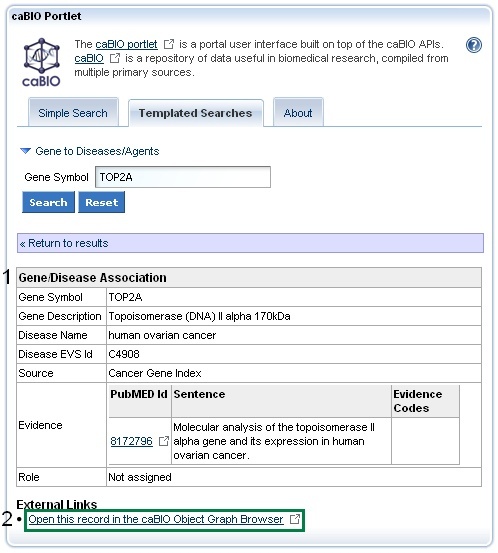
To validate gene-disease associations click on any row in the Gene-Disease Association results table. This will open the concept pair's Gene-Disease Association information. These data include the Gene Term, Gene Symbol, Disease Term, Disease Symbol, Sentence PubMed Identifier, Sentence Evidence, Evidence Code, and Role Code or Detail (1).
Next, click on the Open this record in the caBIO Object Graph Browser at the bottom of the page (2).
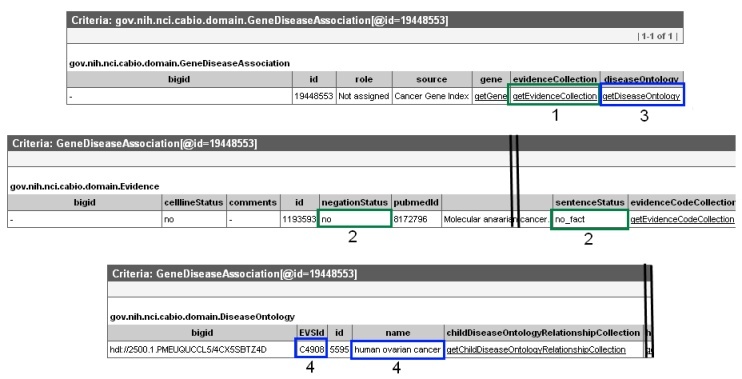
This will open up a the Gene-Disease Assocation object in a new window or tab in your web browser. Click on the getEvidenceCollection link (1, green box) to open the full Evidence record, which contains all of the annotations made by the curator for that particular piece of evidence of a gene-disease association. Gene-disease pairs with true relationships were validated as being true associations by human curators have a sentenceStatus set to "finished" and a negationStatus set to "no" (2, green boxes in the middle panel). If you wish to exclude data from cell lines, for example, the celllineStatus must be be "no." If you want to see some additional information about the disease, navigate back to the Gene-Disease Association object in your browser, and click on getDiseaseOntology (3, blue box in top panel). This will open a Disease Ontology record, in which you may see the name and EVS Identifier for the associated disease (4, blue boxes in bottom panel).
Note
If you accidentally navigate to somewhere else in caBIO by clicking on the wrong link in the Object Viewer and are confused, stop and go back!
Once you have finished reviewing the gene-disease association in the caBIO Object Graph Browser, return to the caBIO Portlet web browser tab or window, and click the Return to Results link in blue to continue evaluating retrieved gene-disease pairs in the results table.
New Gene to Disease/Agent Searches
To search for diseases and compounds/agents associated with other genes, return to your Templated Search results page, if needed, and click the Gene to Diseases/Agents link (1), hit the Reset button (2), enter a new gene symbol (3), and click Search (4).
If you would like to select a new query option, click Return to Templates (below 2 and 4).