Infrastructure for Algorithm Comparisons, Benchmarks, and Challenges in Medical Imaging
Authors: Jayashree Kalpathy-Cramer and Karl Helmer
Introduction
Challenges are being increasingly viewed as a mechanism to foster advances in a number of domains, including healthcare and medicine. The United States Federal Government, as part of the open-government initiative, has underscored the role of challenges as a way to "promote innovation through collaboration and (to) harness the ingenuity of the American Public." Large quantities of publicly available data and cultural changes in the openness of science have now made it possible to use these challenges and crowdsourcing efforts to propel the field forward.
Sites such as Kaggle , Innocentive , and TopCoder are being used increasing in the computer science and data science communities in a range of creative ways. These are being leveraged by commercial entities such as Walmart in finding qualified employees while rewarding participants with monetary prizes as well as less tangible rewards such as public acknowledgement of their efforts for advancing the field.
In the biomedical domain, challenges have been used effectively in bioinformatics as seen by recent crowd-sourced efforts such as Critical Assessment of Protein Structure Prediction (CASP), the CLARITY Challenge for standardizing clinical genome sequencing analysis and reporting and the cancer Genome atlas Pan-cancer analysis Working Group, DREAM Challenges (Dialogue for Reverse Engineering Assessments and Methods), including the prostate challenge currently underway are being used for the assessment of predictive models of disease.
Some of the key advantages of challenges over conventional methods include 1) scientific rigor (sequestering the test data), 2) comparing methods on the same datasets with the same, agreed-upon metrics, 3) allowing computer scientists without access to medical data to test their methods on large clinical datasets, 4) making resources available, such as source code, and 5) bringing together diverse communities (that may traditionally not work together) of imaging and computer scientists, machine learning algorithm developers, software developers, clinicians, and biologists.
However, despite this potential, there are a number of challenges. Medical data is usually governed by privacy and security policies such as HIPPA that make it difficult to share patient data. Patient health records can be very difficult to completely de-identify. Medical imaging data, especially brain MRIs, can be particularly challenging as one could easily reconstruct a recognizable 3D model of the subject.
Crowdsourcing can blur the lines of intellectual property ownership and can make it difficult to translate the algorithms developed in the context of a challenge into a commercial product. A hypothetical example is the development of an algorithm by a researcher at a university for a contest held by a commercial entity with the express purpose of implementing it in a product. Although the researcher who won the contest may have been compensated monetarily, as the IP was developed during her time at the university, the IP is now owned by the University who many not release the rights to the company without further licensing fees.
The infrastructure requirements to both host and participate in some of the "big data" efforts can be monumental. Medical imaging data can be large, historically requiring the shipping of disks to participants. The computing resourcing needed to process these large datasets may be beyond what is available to individual participants. For the organizers, creating the infrastructure that is secure, robust and scalable can require resources beyond the reach of many researchers. These resources included IT manpower support, compute capability, and domain knowledge.
The medical imaging community has conducted a host of challenges at conferences such as MICCAI and SPIE. However, these have typically have been modest in scope (both in terms of data size and number of participants). Medical imaging data poses additional challenges to both participants and organizers. For organizers, ensuring that the data are free of PHI is both critical and non-trivial. Medical data is typically acquired in DICOM format. However, ensuring that a DICOM file is free of PHI requires domain knowledge and specialized software tools. Multimodal imaging data can be extremely large. Imaging formats for pathology images can be proprietary and interoperability between formats can require additional software development efforts. Encouraging non-imaging researchers (e.g. machine-learning scientists) to participate in imaging challenges can be difficult due to the domain knowledge required to convert medical imaging into a set of feature vectors. For participants, access to large compute clusters with computing power, storage space, and bandwidth can prove difficult. Medical imaging data is challenging for non-imaging researchers.
However, it is imperative that the imaging community develops the tools and infrastructure necessary to host these challenges and potentially enlarge the pool of methods by making it more feasible for non-imaging researchers to participate. Resources such as the Cancer Imaging Archive (TCIA) have greatly reduced the burden for sharing medical imaging data within the cancer community and making these data available for use in challenges. Although a number of challenge platforms exist currently, we are not aware of any systems that meet all the requirements necessary to currently host medical imaging challenges.
In this article, we review a few historical imaging challenges. We then list the requirements we believe to be necessary (and nice to have) to support large-scale multimodal imaging challenges. We then review existing systems and develop a matrix of features and tools. Finally, we make some recommendations for developing Medical Imaging Challenge Infrastructure (MedICI), a system to support medical imaging challenges.
Review of Historical Challenges
Challenges have been popular in a number of scientific communities since the 1990s. In the text retrieval community, the Text REtrieval Conference (TREC), co-sponsored by NIST, is an early example of evaluation campaigns where participants work on a common task using data provided by the organizers and evaluated with a common set of metrics. ChaLearn has organized challenges in machine learning since 2013.
We begin with a brief review of a few medical imaging challenges held in the last decade and review their organization and infrastructure requirements. Medical imaging challenges are now a routine aspect of the highly regarded MICCAI annual meeting. Challenges at MICCAI began in 2007 with a liver segmentation and caudate segmentation challenges.
Grand Challenges in Biomedical Image Analysis maintains a fairly updated list of the challenges from the medical imaging community. In a majority of these challenges, the workflow is as described below (adapted from http://grand-challenge.org/Why_Challenges ).
- A task is defined (the output). In our context, this could be segmentation of a lesion or organ, classification of an imaging study as being benign or malignant, prediction of survival, classification of a patients as being a responder or non-responder, pixel/voxel level classification of tissue or tumor grading.
- A set of images are provided (the input). These images are chosen to be of a sufficient size and diversity to reflect the challenges of the clinical problem. Data is typically split up into training and test datasets. The "truth" is made available to the participants for the training data but not the test data. This reduces the risk of overfitting the data and ensures the integrity of the results.
- An evaluation procedure is clearly defined; given the output of an algorithm on a the test images, one or more metrics are computed that measure the performance, usually a reference output is used in this process, but it could also be a visual evaluation of the results by human experts)
- Participants apply their algorithm to all data in the public test dataset provided. They can estimate their performance on the training test.
- Some challenges have an optional leaderboard phase where a subset of the test images is made available to the participants ahead of the final test. Participants can submit their results to the challenge system and have them evaluated or ranked but these are not considered the final standing.
- The reference standard or "ground truth" is defined using methodology clearly described to the participants but is not made publicly available in order to ensure that algorithm results are submitted to the organizers for publication rather than retained privately.
- Final valuation is carried out by the challenge organizers on the test set where the ground truth is sequestered from the participants.
Radiology and Pathology Challenges for Brain Tumor Imaging at MICCAI 2014
MICCAI 2014 held a day-long cluster of event in brain tumor computation including challenges for brain tumor classification and segmentation. The challenge consisted of radiology as well as pathology images. A majority of the images in the training data were from TCIA. Infrastructure support for the radiology portion of the challenges was provided by Bern University and the Virtual Skeleton Database system. The PAIS system support was provided by Stony Brook University for the pathology imaging.
There were 3 sub-challenges within the radiology challenge. The primary goal of the radiology challenge was to perform segmentation from multimodal MRI of brain tumors. T1 (pre- and post-contrast), T2 and FLAIR MRI images were preprocessed (registered and resampled to 1mm isotropic) by the organizers and made available. Ground truth in the form of label maps (4 color –enhancing, necrosis, non-enhancing tumor and edema) were also provided for the training images in .mha format. Additional sub-tasks included longitudinal evaluation of the segmentations for patients who had imaging from multiple time points. Finally, the third subtask was to classify the tumors into one of the three classes (Low Grade II, Low Grade III, and High Grade IV glioblastoma multiforme (GBM)). However, sub-tasks 2 and 3 were primarily pushed out to future years.
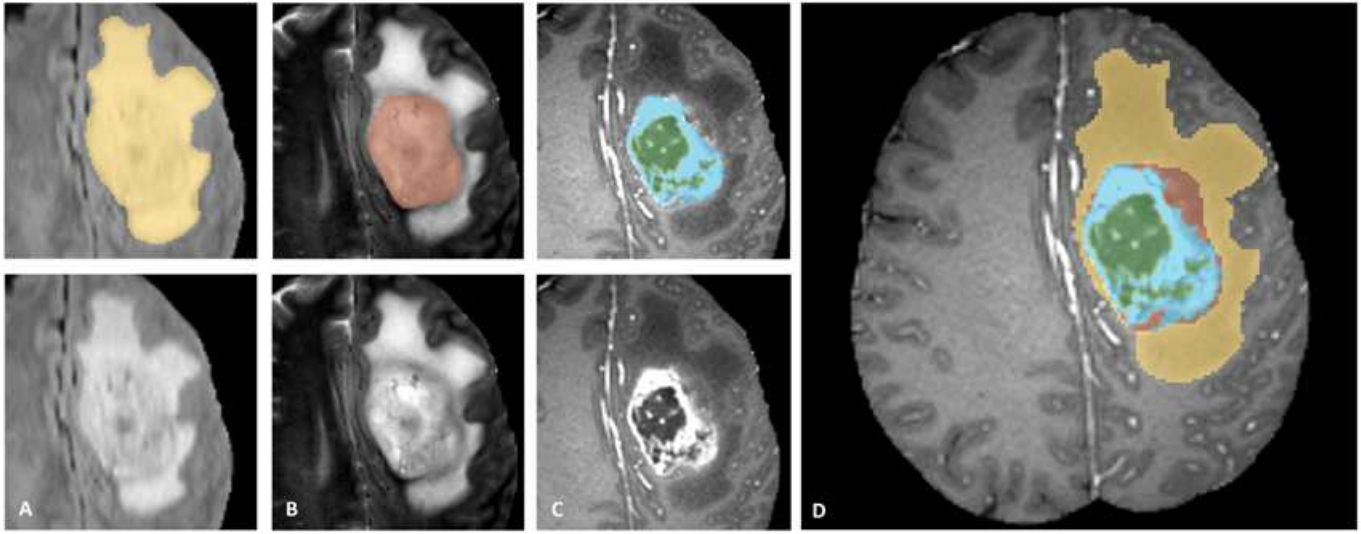
Figure 1. Label maps for the different sub-regions of the tumor used for the BraTS challenge. Manual annotation is performed by expert raters: the whole tumor visible in FLAIR (A), the tumor core visible in T2 (B), the enhancing active tumor visible in T1c (blue), surrounding the cystic/necrotic components of the core (green) (C). The segmentations are combined to generate the final labels (D): edema (yellow), non-enhancing solid core (red), active core (blue), non-solid core (green).
Pathology challenge also had classification and segmentation sub-tasks. The goal for the classification challenge was to classify the image into high grade and low-grade glioma while the goals for the segmentation challenge was to identify areas of necrosis.
The MICCAI-BraTS challenge highlighted a number of findings that mirrored experiences from other domains.
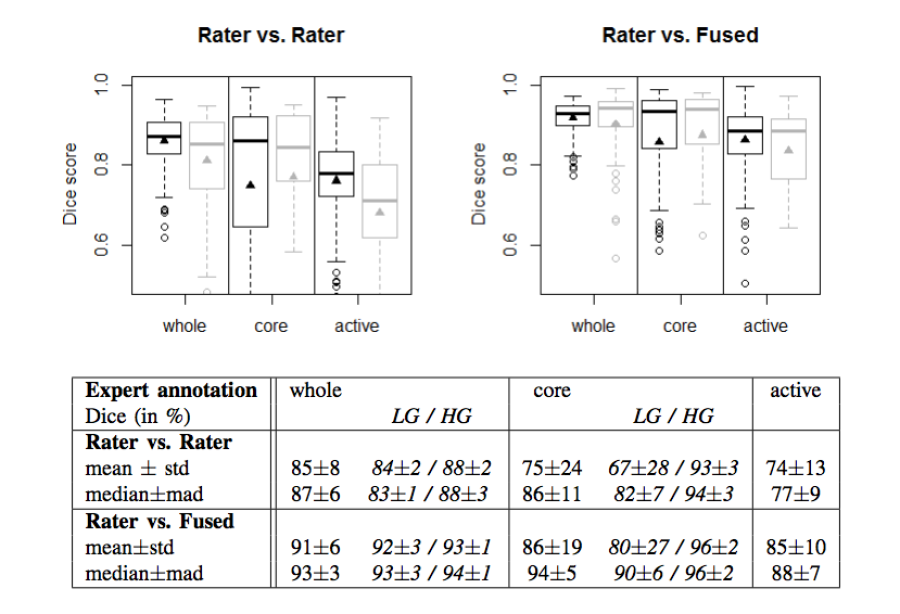
- The agreement between experts is not perfect (~0.8 Dice score).
- The agreement (between experts and between algorithms) is highest for the whole tumor and relatively poor for areas of necrosis and non-enhancing tumor.
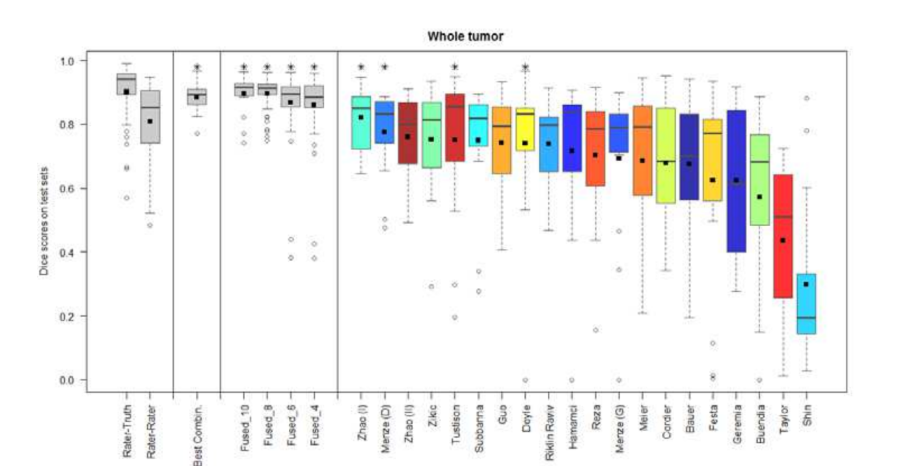
- Combining segmentations created by "best" algorithms created a segmentation that achieves overlap with consensus "expert" labels that approaches inter-rater overlap.
- This approach can be used to automatically create large labeled datasets.
- However, there are cases where this does not work and we still need to validate a subset of images with human experts.
Figure 2. Dice coefficients of inter-rater agreement and of rater vs. fused label maps
Figure 3. Dice coefficients of individual algorithms and fused results indicating improvement with label fusion
More recently, the medical imaging community has begun organizing cloud-based challenges. The VISCERAL project (EU-funded effort) has organized a series of challenges at ISBI, MIICAI and other conferences where the participants in the challenge also share the algorithms and code, in addition to the results.
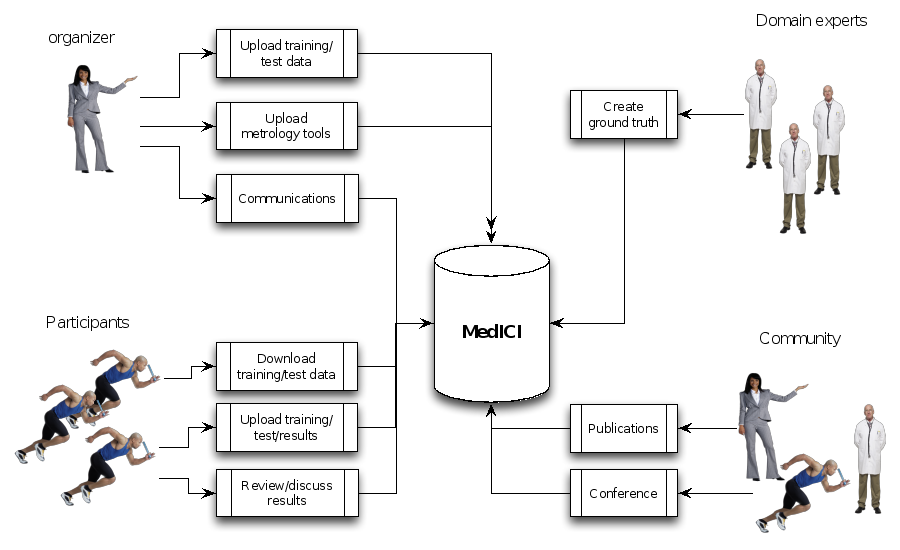
Below is a workflow diagram that describes the various stakeholders in the challenge and their tasks.
Figure 4. Challenge stakeholders and their tasks
Existing Challenge Infrastructure
A number of platforms exist for conducting challenges and crowdsourced efforts. Many of the popular platforms are commercial products, typically offering hosting and organizing services. Challenge organizers work with the company to set up the challenge. In some cases, the challenges are fairly trivial to set up and can be set up with the organizer without much support from the challenge platform company.
Commercial/Hosted
We begin with a brief review a number of popular platforms used for challenges.
These platforms typically charge a hosting fee and offering monetary rewards is pretty common. They have large communities (hundreds of thousands) of registered users and coders and can be a way to introduce the problem to communities outside the core domain expert academic researchers and get solutions that are novel in the domain.
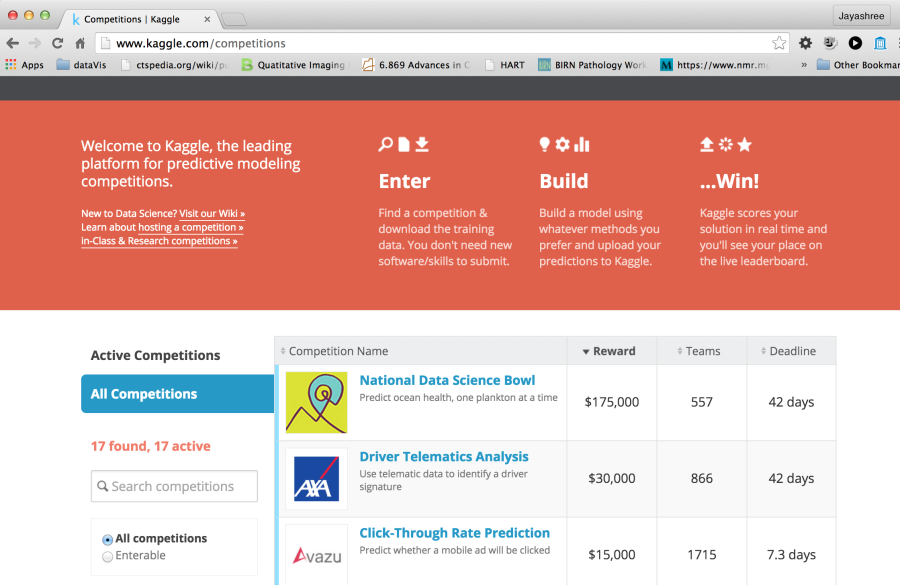
Kaggle is a very popular platform for data science competitions. It is a commercial platform used by companies to pose problems for monetary rewards, jobs and knowledge advancement. There are public and private leaderboards with the test data also being withheld from the participant. Typical hosting costs are reported to be $15,000-20,000 plus additional costs for prizes. However, Kaggle does have a free hosting option to organize challenges for educational purposes. This option is primarily meant to be used by instructors as part of the class curriculum. Kaggle does not provide any support for organizers of Kaggle In Class. There is a 100GB limit on file size. There also appears to be very simple options for scoring. Almost all challenges hosted here appear to be prediction type challenges where results can be submitted as a csv file and the "truth" is also a csv file. It does not appear that imaging-based challenges (such as segmentation challenges) would lend themselves to being hosted on Kaggle In Class without significant effort.
The metrics that Kaggle supports include the following:
Error Metrics for Regression Problems
- Mean Absolute Error
- Weighted Mean Absolute Error
- Root Mean Squared Error
- Root Mean Squared Logarithmic Error
Error Metrics for Classification Problems
- Mean F Score
- Mean Consequential Error
- Mean Average Precision
- Multi Class Log Loss
- Hamming Loss
- Mean Utility
Error Metrics for probability distribution function
- Continuous Ranked Probability Score
Metrics only sensitive to the order
- AUC
- Gini
- Average Among Top P
- Average Precision (column-wise)
- Mean Average Precision (row-wise)
Error Metrics for Ranking Problems
- Normalized Discounted Cumulative Gain@k
- Mean Average Precision@n
Other
- Levenshtein Distance
Other and rarely used:
- Average Precision
- Absolute Error
Figure 5. Portal for Kaggle, a leading website for challenges for data scientists
Topcoder is a similar popular website for software developers, graphic designers and data scientists. In this case, participants typically share their code or designs. They use the Appirio proprietary crowdsourcing development platform, built on Amazon Web Services, Cloud Foundry, Heroku, HTML5, Ruby and Java. A recent computational biology challenge run on Topcoder demonstrated that this crowdsourcing approach produced algorithmic solutions that greatly outperform commonly used algorithms such as BLAST for sequence annotation {Lakhani, 2013 #3789}. This competition was run with a $6000 prize and drew 733 participants (17% of whom submitted code) and the prize-winning algorithms were made available with an open source license.
Challenge Post has been used to organize hackathons, online challenges and other software collaborative activities. In person hackathons are free while the online challenges cost $1500/month (plus other optional charges).
Open Source
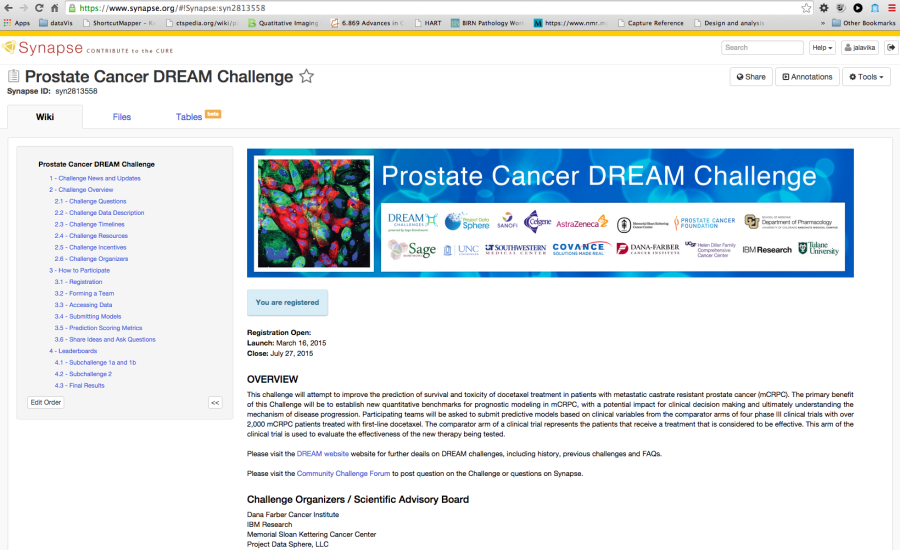
Synapse is both an open source platform and a hosted solution for challenges and collaborative activities created by Sage bionetworks. It has been used for a number of challenges including the DREAM challenge. Synapse allows the sharing of code as well as data. However, the code typically is in R, Python and similar languages. Synapse also has a nice programmatic interface and methods to upload/download data, submit results, create annotations and provenance through R, Python, command line and Java. These options can be configured for the different challenges. Content in Synapse is referenced by unique Synapse IDs. The three basic types of Synapse objects include projects, folders and files. These can be accessed through the web interface or through programmatic APIs. Experience and support for running image analysis code within Synapse is limited.
Figure 6. Portal for the Synapse platform
Figure 7. Example Challenge hosted in Synapse
COMIC framework is an open-source platform that facilitates the creation of challenges and has been used to host a number of medical imaging challenges. The Consortium for Open Medical Image Computing (COMIC) platform, built using Python/Django was created and is maintained by a consortium of five European medical image analysis groups including Radboud University, Erasmus, and UCL. They also offer a hosted site, with the hardware located at Fraunhofer MEVIS in Bremen, Germany. The current framework allows participants to create a website, add pages including wikis, create participant registrations, methods for organizers to upload data and participants to download data (for instance through Dropbox). However, the platform including ways to visualize medical data and results is still under development as are options to share algorithms and perform challenges in the cloud.
The main steps to create a new challenge are:
- Create a project
- Add pages
- Making uploaded files available for download
- Allowing others to register for your project
- Make your project appear in the projects overview
- Allow file uploads
- Including content from files on a page
- Allow others to edit the project
- Changing colors and other styling
- Project data folder
- Page permissions
However, at this time, there is limited support for automatic evaluation of submitted results, results presentation, native support for medical images although many of these features are planned.
The HubZero is an open source platform developed for scientific collaboration. It has been used heavily in a number of communities including nanoscience, earthquake engineering, molecular diagnostics and others. A version focused on cancer informatics can is hosted at nciphub.org. nciphub shares a lot of features with the Synapse platform. It allows user management and role-based access. Users can create groups that share common interest and collaborate within these groups. Files can be shared within projects. Other features include wiki, calendars, creating and sharing resources such as presentations, multimedia and even tools. Most common tools found on the various hubs are those based on simulations. Although nciphub has limited native support of medical imaging, libraries to handle medical images can be configured to work in the hub. Members of the Quantitative Imaging Network (QIN) are exploring the use of nciphub for challenges, especially for the communication and data sharing.
CodaLab is an open-source project that originated at Microsoft Research that was expressly created for hosting challenges and supporting reproducible research. The OuterCurve Foundation currently maintains it. Challenge organizers can easily set up challenges by creating a competition bundle that consists of data as well as evaluate tools. As part of the configuration files, the number of phases and duration (e.g. training, leaderboard, test) can be set up by the organizer. The evaluation program can be written in any language. Participants can upload results and get immediate feedback. The currently available version of CodaLab comes with scoring algorithms for image segmentation evaluation Organizers can extend the presentation of results to allow drilling down into the results with tables and charts. CodaLab currently uses the Azure platform although, in theory, it should be possible to deploy on other servers without a great deal of effort. CodaLab is also developing support for worksheets. These are resources to support reproducible research and for collaboration. Using these, researchers have compared a number of open source NLP tools on different public datasets . As this technology continues to be developed, researchers will be able to quickly compare the performance of different algorithms on a range of datasets in the "cloud" by leveraging Azure technology.
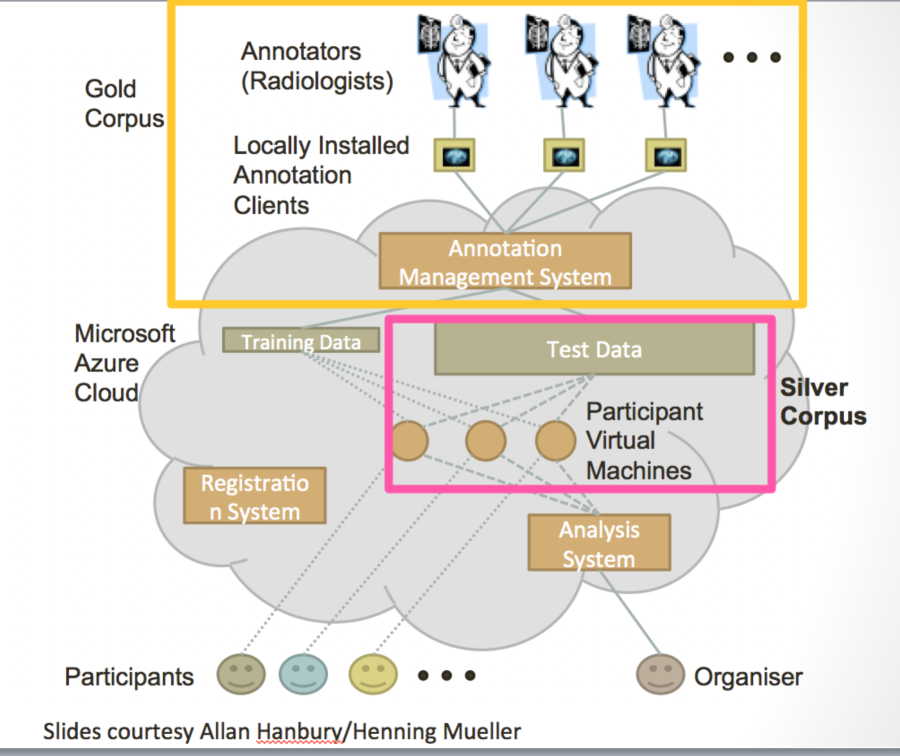
Visual Concept Extraction Challenge in Radiology (VISCERAL) is a large EU funded project to develop cloud-based challenge infrastructure. This open source platform, based on the Azure platform as described below, facilitates cloud-based challenges where the participants upload their algorithms rather than downloaded data and uploading algorithm output. This platform has been used for 4 medical imaging challenges at MICCAI and ISBI. Participants are provided virtual machines with access to the training data where they can deploy, configure and validate their algorithms. Once the training phase is completed, the virtual machines are then handed over to the organizers. The organizers can then run the algorithms on the test data. This feature, where the organizers and not the participants run the algorithms on the test data, is a unique to the VISCERAL system. This has a number of advantages in that the participants are never provided access to the test data, which reduces the risk of overfitting. Furthermore, it allows private data to be used for the testing phase and promotes unplanned dissemination of secure data. Finally, it supports the notion of reproducible research as the algorithms can always be rerun if the virtual machines are saved. Participants are allowed the share either source code or executables thus allowing both open and closed source algorithms to compete in the same venue.
Figure 8. Schematic diagrams of the VISCERAL system for cloud-based challenges
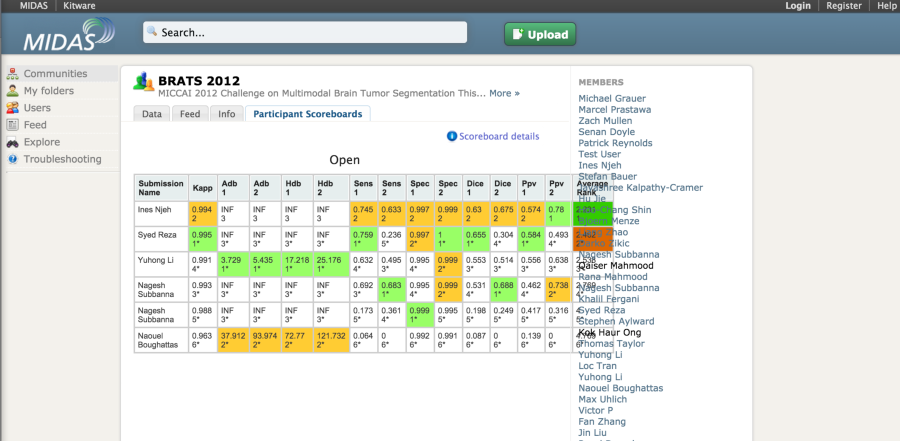
The MIDAS platform has been used to host a couple of imaging challenges. A special module is available to host challenges. The developers of the platform also made available the COVALIC evaluation tool for segmentation challenges with the following metrics: Average distance of boundary surfaces, 95th percentile Hausdorff distance of boundary surfaces, Dice overlap, Cohen's kappa, Sensitivity, Specificity, Positive Predictive Value.
Once the participants have uploaded their submissions, the leaderboard updates the scores automatically.
A new version of the platform appears to be in development. This system (COVALIC) is built on the TangeloHub platform--an open source data and analytics platform made up of three major components: Tangelo, Girder, and Romanesco.
Matrix of Features and Frameworks (1 -5)
Below is a table that rates the relative merits of the most relevant frameworks that we evaluated. The scale is 1-5 where 1 indicates excellent support for the feature while 5 indicates that that feature is not currently part of the system or there is limited support. We have included Kaggle as a representative "paid" framework for comparison. The other five frameworks are the open-source frameworks that were seriously considered in this comparison.
| Kaggle | Synapse | HubZero (challenges/projects) | COMIC | VISCERAL | CodaLab | |
|---|---|---|---|---|---|---|
| Ease of setting up new challenge | 2/4 (if new metrics need to be used) | 2 | 2/5 | 2 | 3 | 1 |
Cost (own server/hosting options) | $10-$25k/challenge | Free/hosted | Free/hosted | Free/hosted | Free/Azure costs | Free/hosted |
License | Commercial | OS | OS | OS | OS | OS |
Ease of extensibility | 5 | 4 | 4 | 2 | 3 | 2 |
Cloud support for algorithms | 4 | 3 | 3 | 4 | 1 | 3 |
Maturity | 1 | 1 | 1/5 | 3 | 4 | 3 |
Flexibility |
|
|
|
|
|
|
Number of users | 1 | 1 | 1/5 | 3 | 3 | 3 |
Types of challenges | 1 | 1 | 1 | 3 | 1 | 1 |
Native imaging support | No | No | No | Yes | Limited | No |
API to access data, code | 5 | 1 | 3 | 4 | 4 | 4 |
Components of Challenge Infrastructure
We describe below the various components of challenge infrastructure that would be necessary to host joint radiology/pathology challenges.
The web portal is the single point of entry for the participants. Historically, this would have information about the challenge, potentially host the data and provide a submission site for the user to upload results. The challenge organizer could also provide the results of the challenge at this page. Many challenges have wikis and announcement pages as well as forums. A good example of active discussion forums can be found at the Kaggle . Most systems have backend systems (typically a relational database) for managing data and users. These allow registered used to access perhaps the training data and ground truth, the test data but not the ground truth.
Challenge systems tailored for radiology and pathology also have specialized tools for handling these specialized data types and for creating and management of annotations and ground truth. Challenge systems also need modules for scoring and evaluation of the submissions. Finally, it is important to present the results back to the participants. Often these are presented in an ordered fashion with "winners at the top of the list.
Conclusions: Trade-Offs and Recommendations
Challenges have a very important role in moving science forward. In this document, we reviewed some of the more popular platforms to host challenges and compare some of the key aspects of these platforms. We believe that challenge infrastructure should be modular, flexible, extensible and user friendly. Requirements for this platform included support for radiology and pathology challenges. We were primarily seeking an open-source option. Although no single platform met all the requirements for our purposes, we were seeking solutions that could be extended easily and potentially had good interfaces that could be used to tie components together. We were envisioning potentially one solution for the more general aspects of challenge management (user and organizer management, data download, results upload, evaluation, results display), while adding other modules that are more specific for radiology and pathology imaging. A modular solution would allow us to switch out components as technologies mature or new technologies emerge.
Although Kaggle, Innocentive and Topcoder all have platforms that have been used extensively for a really wide range of challenges, these were excluded from further consideration (and from the above table) as platforms since they are not open source and cannot be modified.
The Synapse platform has been used for a number of challenges including ones where the source code has been shared. However, it has not been used to conduct medical imaging challenges. Also, the code base for the platform is quite extensive and would potentially be quite difficult to modify and contribute to, for outside investigators.
For the basic challenge infrastructure for classic challenges where the participants download data and upload results, MIDAS (and potentially the replacement COVALIC), the COMIC framework and CodaLab are all viable options. They are all open source and have been used to conduct a few challenges (2-20). The MIDAS challenge/COVALIC platforms appear to be in the midst of a complete overhaul and thus were not quite ready for a comprehensive evaluation at this point. The COMIC framework and CodaLab code base are of more manageable size and we believe that we would be able to extend them quite easily. Organizers from the community that were not developers of the platform have used both platforms without support from the platform team, a critical criteria for us.
The COMIC framework, admittedly, is still under development. "The tools we offer include an easy way to create a site, add and edit pages like a wiki, registration mechanisms for participants, secure ways for organizers to upload the challenge data and for participants to download it, for participants to upload results, ways to tabulate, sort and visualize the results, and much more. Much of this is still under development, such as ways to visualize medical data and results of algorithms interactively, directly in your browser, to upload evaluation code that processes new results immediately "in the cloud", and to upload the algorithms themselves if the developer is willing to share these." They use Dropbox for image sharing.
Based on the time frame of this project and the current state of development of the platforms that we evaluated, we chose the CodaLab platform for the first iteration of MedICI as we believed that it offered the greatest compromise between current features, ease of use, flexibility and extensibility. However, we will re-evaluate this decision in 3 months and believe that we could very easily port any code we develop to other platforms. For the radiology annotations and metadata management, we chose ePad while we chose caMicroscope for the pathology annotation and metadata management.
Reference
- Shi S, Pei J, Sadreyev RI, Kinch LN, Majumdar I, Tong J, Cheng H, Kim BH, Grishin NV. Analysis of CASP8 targets, predictions and assessment methods. Database : the journal of biological databases and curation. 2009;2009:bap003. doi: 10.1093/database/bap003. PubMed PMID: 20157476; PubMed Central PMCID: PMC2794793.
- Brownstein CA, Beggs AH, Homer N, Merriman B, Yu TW, Flannery KC, DeChene ET, Towne MC, Savage SK, Price EN, Holm IA, Luquette LJ, Lyon E, Majzoub J, Neupert P, McCallie D, Jr., Szolovits P, Willard HF, Mendelsohn NJ, Temme R, Finkel RS, Yum SW, Medne L, Sunyaev SR, Adzhubey I, Cassa CA, de Bakker PI, Duzkale H, Dworzynski P, Fairbrother W, Francioli L, Funke BH, Giovanni MA, Handsaker RE, Lage K, Lebo MS, Lek M, Leshchiner I, MacArthur DG, McLaughlin HM, Murray MF, Pers TH, Polak PP, Raychaudhuri S, Rehm HL, Soemedi R, Stitziel NO, Vestecka S, Supper J, Gugenmus C, Klocke B, Hahn A, Schubach M, Menzel M, Biskup S, Freisinger P, Deng M, Braun M, Perner S, Smith RJ, Andorf JL, Huang J, Ryckman K, Sheffield VC, Stone EM, Bair T, Black-Ziegelbein EA, Braun TA, Darbro B, DeLuca AP, Kolbe DL, Scheetz TE, Shearer AE, Sompallae R, Wang K, Bassuk AG, Edens E, Mathews K, Moore SA, Shchelochkov OA, Trapane P, Bossler A, Campbell CA, Heusel JW, Kwitek A, Maga T, Panzer K, Wassink T, Van Daele D, Azaiez H, Booth K, Meyer N, Segal MM, Williams MS, Tromp G, White P, Corsmeier D, Fitzgerald-Butt S, Herman G, Lamb-Thrush D, McBride KL, Newsom D, Pierson CR, Rakowsky AT, Maver A, Lovrecic L, Palandacic A, Peterlin B, Torkamani A, Wedell A, Huss M, Alexeyenko A, Lindvall JM, Magnusson M, Nilsson D, Stranneheim H, Taylan F, Gilissen C, Hoischen A, van Bon B, Yntema H, Nelen M, Zhang W, Sager J, Zhang L, Blair K, Kural D, Cariaso M, Lennon GG, Javed A, Agrawal S, Ng PC, Sandhu KS, Krishna S, Veeramachaneni V, Isakov O, Halperin E, Friedman E, Shomron N, Glusman G, Roach JC, Caballero J, Cox HC, Mauldin D, Ament SA, Rowen L, Richards DR, San Lucas FA, Gonzalez-Garay ML, Caskey CT, Bai Y, Huang Y, Fang F, Zhang Y, Wang Z, Barrera J, Garcia-Lobo JM, Gonzalez-Lamuno D, Llorca J, Rodriguez MC, Varela I, Reese MG, De La Vega FM, Kiruluta E, Cargill M, Hart RK, Sorenson JM, Lyon GJ, Stevenson DA, Bray BE, Moore BM, Eilbeck K, Yandell M, Zhao H, Hou L, Chen X, Yan X, Chen M, Li C, Yang C, Gunel M, Li P, Kong Y, Alexander AC, Albertyn ZI, Boycott KM, Bulman DE, Gordon PM, Innes AM, Knoppers BM, Majewski J, Marshall CR, Parboosingh JS, Sawyer SL, Samuels ME, Schwartzentruber J, Kohane IS, Margulies DM. An international effort towards developing standards for best practices in analysis, interpretation and reporting of clinical genome sequencing results in the CLARITY Challenge. Genome biology. 2014;15(3):R53. doi: 10.1186/gb-2014-15-3-r53. PubMed PMID: 24667040; PubMed Central PMCID: PMC4073084.
- Omberg L, Ellrott K, Yuan Y, Kandoth C, Wong C, Kellen MR, Friend SH, Stuart J, Liang H, Margolin AA. Enabling transparent and collaborative computational analysis of 12 tumor types within The Cancer Genome Atlas. Nature genetics. 2013;45(10):1121-6. doi: 10.1038/ng.2761. PubMed PMID: 24071850; PubMed Central PMCID: PMC3950337.
- Abdallah K, Hugh-Jones C, Norman T, Friend S, Stolovitzky G. The Prostate Cancer DREAM Challenge: A Community-Wide Effort to Use Open Clinical Trial Data for the Quantitative Prediction of Outcomes in Metastatic Prostate Cancer. The oncologist. 2015. doi: 10.1634/theoncologist.2015-0054. PubMed PMID: 25777346.
- Jarchum I, Jones S. DREAMing of benchmarks. Nat Biotechnol. 2015;33(1):49-50. doi: 10.1038/nbt.3115. PubMed PMID: 25574639.