- Documentation Main Page
- Creation of the Cancer Gene Index
- Data, Metadata, and Annotations
- Cancer Gene Index Gene-Disease and Gene-Compound XML Documents
- caBIO APIs
- Cancer Gene Index Shared Parsed Data and Code
- caBIO Portlet Templated Searches
- caBIO Home Page
- caBIO iPhone Application
- caBIO Portlet Simple Searches
- Glossary
- Credits and Resources
To Print the Guide
We recommend you print one wiki page of the guide at a time. To do this, click the printer icon at the top right of the page; then from the browser File menu, choose Print. Printing multiple pages at one time is more complex. For instructions, refer to Printing multiple pages .
Having Trouble Reading the Text?
Resizing the text for any web page is easy. For information on how to do this in your web browser, refer to this W3C tutorial
Cancer Gene Index XML Overview
The Cancer Gene Index is available as two ZIP files that contain the data from the Gene-Disease and Gene-Compound Databases. The Cancer Gene Index Gene-Disease and Gene-Compound "Databases" each include an XML document and an accompanying DTD, CancerIndex_disease_XML.dtd and CancerIndex_compound_XML.dtd, respectively. You may freely download the Gene-Disease file and Gene-Compound file from the Cancer Gene Index website.
XML System Requirements
The Cancer Gene Index Gene-Disease and Gene-Compound data sets require at least 720 MB of available hard drive space.
You can use these documents to uncover fact-based associations between genes and diseases or genes and compounds. In addition, you can evaluate the evidence from which these associations were extracted (that is, the sentences from MEDLINE abstracts) and the codes and details that describe these associations. Other annotations, such as sentence status flags, negation indicators, cell line indicators, and organism data are information-rich and may be used to limit queries of the data, for example. Additional information is available in the Using the Cancer Gene Index subsection of this document.
Cancer Gene Index DTDs
Descriptions of the Gene-Disease and Gene-Compound DTD elements are provided below.
Description of the Cancer Gene Index Gene-Disease DTD Elements
Gene-Disease DTD Element | Description |
|---|---|
<!ELEMENT GeneEntryCollection (GeneEntry+)> | A collection of all gene, disease, evidence, and annotation information associated with a gene concept |
<!ELEMENT GeneEntry (HUGOGeneSymbol, GeneAliasCollection, SequenceIdentificationCollection, GeneStatusFlag, Sentence*)> | All information associated with a particular gene concept |
<!ELEMENT HUGOGeneSymbol (#PCDATA)> | HUGO Gene Symbol for the gene concept |
<!ELEMENT GeneAliasCollection (GeneAlias+)> | A collection of acronyms, synonyms, alternate spellings, and other aliases for the gene concept |
<!ELEMENT GeneAlias (#PCDATA)> | A specific synonym, alternate spelling, or other alias for the gene concept |
<!ELEMENT SequenceIdentificationCollection (HgncID, LocusLinkID, GenbankAccession, RefSeqID, UniProtID)> | A collection of standard identifiers for the gene concept |
<!ELEMENT HgncID (#PCDATA)> | HGNC Identifier for the gene concept A |
<!ELEMENT LocusLinkID (#PCDATA)> | LocusLink Identifier for the gene concept A |
<!ELEMENT GenbankAccession (#PCDATA)> | Genbank Accession Number for the gene concept B |
<!ELEMENT RefSeqID (#PCDATA)> | RefSeq Identifier for the gene concept C |
<!ELEMENT UniProtID (#PCDATA)> | UniProt Identifier corresponding to the gene concept C |
<!ELEMENT GeneStatusFlag (#PCDATA)> | The status of the gene set by a human curator |
<!ELEMENT Sentence (GeneData, DiseaseData, Statement, PubMedID, Organism, NegationIndicator, CellineIndicator, Comments?, EvidenceCode*, Roles*, SentenceStatusFlag)> | Data and annotations for the extracted sentence for gene-disease concept pairs |
<!ELEMENT GeneData (MatchedGeneTerm, NCIGeneConceptCode)> | The Gene Term and EVS Gene Concept Code Identifier for the gene concept of the gene-disease concept pair |
<!ELEMENT MatchedGeneTerm (#PCDATA)> | Matched term of the gene concept |
<!ELEMENT NCIGeneConceptCode (#PCDATA)> | Gene Concept Code corresponding to the Matched Gene Term B |
<!ELEMENT DiseaseData (MatchedDiseaseTerm, NCIDiseaseConceptCode)> | Disease Term and EVS Concept Code Identifier for the disease concept of the gene-disease concept pair |
<!ELEMENT MatchedDiseaseTerm (#PCDATA)> | NCI Thesaurus Matched Disease Term for the disease concept |
<!ELEMENT NCIDiseaseConceptCode (#PCDATA)> | EVS Disease Concept Code corresponding to the Matched Disease Term |
<!ELEMENT Statement (#PCDATA)> | Sentence statement containing the evidence of the gene-disease association |
<!ELEMENT PubMedID (#PCDATA)> | PubMed Identifier for the abstract from which the evidence was extracted |
<!ELEMENT Organism (#PCDATA)> | Organism from which the data were collected |
<!ELEMENT NegationIndicator (#PCDATA)> | Whether the findings of a gene-disease association within a sentence were negative |
<!ELEMENT CellineIndicator (#PCDATA)> | Whether the data were collected from a cell line |
<!ELEMENT Comments (#PCDATA)> | Comments made by expert curators |
<!ELEMENT EvidenceCode (#PCDATA)> | Evidence Code |
<!ELEMENT Roles (PrimaryNCIRoleCode*, OtherRole*)> | Role Code and Role Detail for the gene-disease concept pair |
<!ELEMENT PrimaryNCIRoleCode (#PCDATA)> | Role Code and Role Detail for the gene-disease concept pair |
<!ELEMENT OtherRole (#PCDATA)> | Role Detail |
<!ELEMENT SentenceStatusFlag (#PCDATA)> | Sentence Status Flag set by the expert curators |
A Some of the genes in the gene-disease concept pairs are not included in the HGNC or LocusLink and, thus, the text contents for these elements will be "0"
B The text contents for this element are " "
C RefSeq and UniProt Identifiers were taken from HGNC, which does not include all gene concepts included in the Cancer Gene Index. For the affected genes, the element's text contents will be "-"
Description of the Cancer Gene Index Gene-Compound DTD Elements
Gene-Disease DTD Element | Description |
|---|---|
<!ELEMENT GeneEntryCollection (GeneEntry+)> | A collection of all gene, compound, evidence, and annotation information associated with a gene concept |
<!ELEMENT GeneEntry (HUGOGeneSymbol, GeneAliasCollection, SequenceIdentificationCollection, GeneStatusFlag, Sentence*)> | All information associated with a particular gene concept |
<!ELEMENT HUGOGeneSymbol (#PCDATA)> | HUGO Gene Symbol for the gene concept |
<!ELEMENT GeneAliasCollection (GeneAlias+)> | A collection of acronyms, synonyms, alternate spellings, and other aliases for the gene concept |
<!ELEMENT GeneAlias (#PCDATA)> | A specific synonym, alternate spelling, or other alias for the gene concept |
<!ELEMENT SequenceIdentificationCollection (HgncID, LocusLinkID, GenbankAccession, RefSeqID, UniProtID)> | A collection of standard identifiers for the gene concept |
<!ELEMENT HgncID (#PCDATA)> | HGNC Identifier for the gene concept A |
<!ELEMENT LocusLinkID (#PCDATA)> | LocusLink Identifier for the gene concept A |
<!ELEMENT GenbankAccession (#PCDATA)> | Genbank Accession Number for the gene concept B |
<!ELEMENT RefSeqID (#PCDATA)> | RefSeq Identifier for the gene concept C |
<!ELEMENT UniProtID (#PCDATA)> | UniProt Identifier corresponding to the gene concept C |
<!ELEMENT GeneStatusFlag (#PCDATA)> | The status of the gene set by a human curator |
<!ELEMENT Sentence (GeneData, DiseaseData, Statement, PubMedID, Organism, NegationIndicator, CellineIndicator, Comments?, EvidenceCode*, Roles*, SentenceStatusFlag)> | Data and annotations for the extracted sentence for gene-compound concept pairs |
<!ELEMENT GeneData (MatchedGeneTerm, NCIGeneConceptCode)> | The Gene Term and EVS Gene Concept Code Identifier for the gene concept of the gene-compound concept pair |
<!ELEMENT MatchedGeneTerm (#PCDATA)> | Matched term of the gene concept |
<!ELEMENT NCIGeneConceptCode (#PCDATA)> | Gene Concept Code corresponding to the Matched Gene Term B |
<!ELEMENT DrugData (MatchedDrugTerm, NCIDrugConceptCode)> | Compound Term and EVS Concept Code Identifier for the compound concept of the gene-compound concept pair |
<!ELEMENT MatchedDrugTerm (#PCDATA)> | NCI Thesaurus Matched Compound Term for the compound concept |
<!ELEMENT NCIDrugConceptCode (#PCDATA)> | EVS Compound Concept Code corresponding to the Matched Compound Term |
<!ELEMENT Statement (#PCDATA)> | Sentence statement containing the evidence of the gene-compound association |
<!ELEMENT PubMedID (#PCDATA)> | PubMed Identifier for the abstract from which the evidence was extracted |
<!ELEMENT Organism (#PCDATA)> | Organism from which the data were collected |
<!ELEMENT NegationIndicator (#PCDATA)> | Whether the findings of a gene-compound association within a sentence were negative |
<!ELEMENT CellineIndicator (#PCDATA)> | Whether the data were collected from a cell line |
<!ELEMENT Comments (#PCDATA)> | Comments made by expert curators |
<!ELEMENT EvidenceCode (#PCDATA)> | Evidence Code |
<!ELEMENT Roles (PrimaryNCIRoleCode*, OtherRole*)> | Role Code and Role Detail for the gene-compound concept pair |
<!ELEMENT PrimaryNCIRoleCode (#PCDATA)> | Role Code and Role Detail for the gene-compound concept pair |
<!ELEMENT OtherRole (#PCDATA)> | Role Detail |
<!ELEMENT SentenceStatusFlag (#PCDATA)> | Sentence Status Flag set by the expert curators |
A Some of the genes in the gene-compound concept pairs are not included in the HGNC or LocusLink and, thus, the text contents for these elements will be "0"
B The text contents for this element are " "
C RefSeq and UniProt Identifiers were taken from HGNC, which does not include all gene concepts included in the Cancer Gene Index. For the affected genes, the element's text contents will be "-"
Additional DTD Information
The Gene-Disease and Gene-Compound DTD elements include meaningful parenthetical information and special characters. Consider the following examples:
<!ELEMENT GeneEntry (HUGOGeneSymbol, GeneAliasCollection, SequenceIdentificationCollection, GeneStatusFlag, Sentence<!ELEMENT HUGOGeneSymbol (#PCDATA)><!ELEMENT GeneAliasCollection (GeneAlias+)><!ELEMENT GeneAlias (#PCDATA)><!ELEMENT Sentence (GeneData, DiseaseData, Statement, PubMedID, Organism, NegationIndicator, CellineIndicator, Comments?, EvidenceCode*, Roles*, SentenceStatusFlag)>
!ELEMENT GeneEntry defines that the GeneEntry element contains five child elements: HUGOGeneSymbol, GeneAliasCollection, SequenceIdentificationCollection, GeneStatusFlag, Sentence*
!ELEMENT HUGOGeneSymbol defines the HUGOGeneSymbol element to be of type #PCDATA
!ELEMENT GeneAliasCollection defines the GeneAliasCollection element to be of type GeneAlias+
!ELEMENT GeneAlias defines the GeneAlias element to be of type #PCDATA
!ELEMENT Sentence defines the Sentence element contains eleven elements: GeneData, DiseaseData, Statement, PubMedID, Organism, NegationIndicator, CellineIndicator, Comments?, EvidenceCode*, Roles*, SentenceStatusFlag
#PCDATA stands for Parsed Character data. Declarations of element type #PCDATA mean that XML Parsers will parse the text contents found between the start and end tags of an XML element that correspond to this DTD element.
Cancer Gene Index elements not only contain child elements and text elements, but also information about the presence of child elements and the number of times a particular element can recur. Elements with one or more child elements declare the name(s) of the child elements as comma-separated lists inside parentheses. Examples of Cancer Gene Index elements with multiple child elements are given above in 1 and 5.
Note
Child elements appear in the same order in the XML documents as the DTDs, and they can themselves have one or more children, as described below.
Special characters (for example, +, *, ?) appended to the name of a child element describe the expected number of occurrences of element. The + character in example 3 above declares that the child element GeneAlias must occur one or more times inside the GeneAliasCollection element. The * character in examples 1 and 5 above declares that the child element Sentence, EvidenceCode, and Roles can occur zero or more times inside the GeneEntry and Sentence elements. The ? character in example 5 above declares that the child element Comments can be absent or occur one time inside the Sentence element.
Parsing the Cancer Gene Index XML
Many free XML parsers exist, as do parsing modules or libraries for a variety of common programming languages, that will quickly divide the Gene-Disease and Gene-Compound XML documents into their component data. Parsed data can be stored in a database or other data management application and be computed against. Alternatively, you may prefer to write code that recursively loops through the XML and extracts the information that you desire. As end users parse the Cancer Gene Index data into various formats (for example, database dumps or tab-delimited text files) or create code to walk through the XML, they are strongly encouraged to make these versions and the code available by posting them to the Cancer Gene Index User Community Parsed Data and Code web page.
Using the Cancer Gene Index Data
The following subsections provide information and tips to maximize your use of the Cancer Gene Index XML files.
Refining Your Searches with Flags and Indicators
You can use the Cancer Gene Index to discover associations between genes and diseases or genes and compounds. These associations were derived from the literature using a sophisticated automated process, and thus not all of the extracted gene-disease or gene-compound concept pair associations were found to be factual by expert human curators in subsequent validation steps.
Tip
If you would like to restrict your queries of the Cancer Gene Index data sets to only those concept pairs that were validated as being truly associated, filter out information where the SentenceStatusFlag is "no_fact" or "unclear" and where the NegationIndicator is "yes."
You also, of course, can take advantage of other annotations such as CelllineIndicator or Organism names. For information about the status flags, indicators, and other annotations within the XML documents, refer to the Cancer Gene Index Data, Metadata, and Annotations wiki page.
Codes and Details
The expert curators also set Evidence Codes, Role Codes, and Role Details for each concept pair. Evidence codes (EvidenceCodes) qualify the assertions of a gene-disease or gene-compound association made in the sentence and provide information on how the these assertions were made. Role Codes (PrimaryNCIRoleCode) and Role Details (OtherRole) describe the semantic associations between gene and either a disease or compound term. Whereas the Evidence Codes describe how the association was inferred or the type of experiment upon which the inference was made, Role Codes and Role Details give information about the actual gene-disease or gene-compound association.
For information about the meaning of the codes, details, and other data and annotations within the XML documents, refer to the Cancer Gene Index Data, Metadata, and Annotations wiki page.
Gene, Disease, and Compound Ontologies
Although the NCI Thesaurus provides compound, disease, and gene ontological information and was used to create the data resource, this information is not easily deduced from data in the Cancer Gene Index, itself.
Should you wish to search for parent, sister, and child concepts, it is possible to trace back to the hierarchical disease, compound, and gene terms with the NCI Thesaurus GUI or the Enterprise Vocabulary Services (EVS) API. To use these tools, you will need the NCI Thesaurus concept terms (for example, from MatchedGeneTerm or MatchedDrugTerm) or NCI Thesaurus Concept Code (for example, from NCIDiseaseConceptCode or NCIDrugConceptCode).
Tip
The NCI Thesaurus Concept Code for a gene, disease, or compound term is also its EVS Identifier.
Using the NCI Thesaurus to Find Parent/Child Concepts
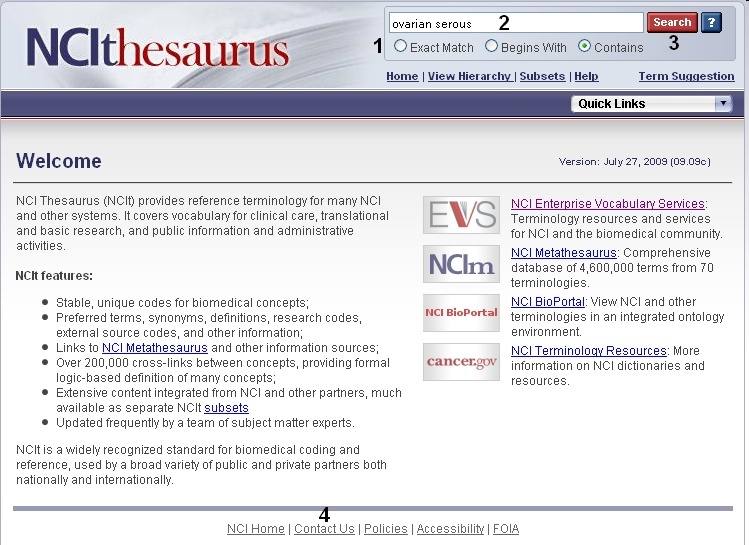
To view disease, compound, and gene ontologies, open a new browser tab or window and navigate to the NCI Thesaurus web page, enter in your gene symbol, matched term, or NCI Thesaurus concept code (2, "ovarian serous adenocarcinoma"), and click the Search button (3). If you need help finding your gene, disease, or compound term, click the Contact Us link at the bottom of the page (4).
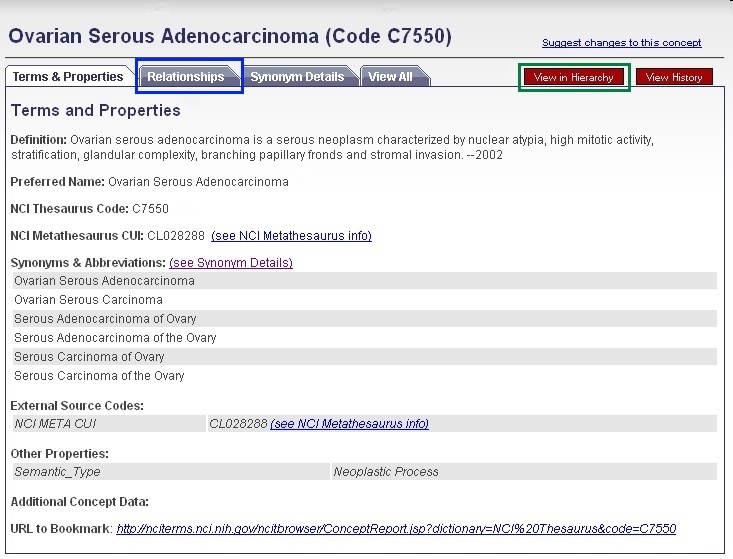
You may view parent and child terms for any disease term by clicking the Relationships tab (selected in figure). For example, "ovarian serous adenocarcinoma" has the children "ovarian serous cystadenocarcinoma" and "ovarian serous papillary adenocarcinoma" and the parent terms "malignant ovarian serous tumor," "ovarian adenocarcinoma," and "serous adenocarcinoma." Alternatively, if you would like to view where your term fits in the entire disease hierarchy, click the red View in Hierarchy button (selected in figure).