- Documentation Main Page
- Creation of the Cancer Gene Index
- Data, Metadata, and Annotations
- Cancer Gene Index Gene-Disease and Gene-Compound XML Documents
- caBIO APIs
- Cancer Gene Index Shared Parsed Data and Code
- caBIO Portlet Templated Searches
- caBIO Home Page
- caBIO iPhone Application
- caBIO Portlet Simple Searches
- Glossary
- Credits and Resources
To Print the Guide
We recommend you print one wiki page of the guide at a time. To do this, click the printer icon at the top right of the page; then from the browser File menu, choose Print. Printing multiple pages at one time is more complex. For instructions, refer to How do I print multiple pages?.
Having Trouble Reading the Text?
Resizing the text for any web page is easy. For information on how to do this in your web browser, refer to this W3C tutorial .
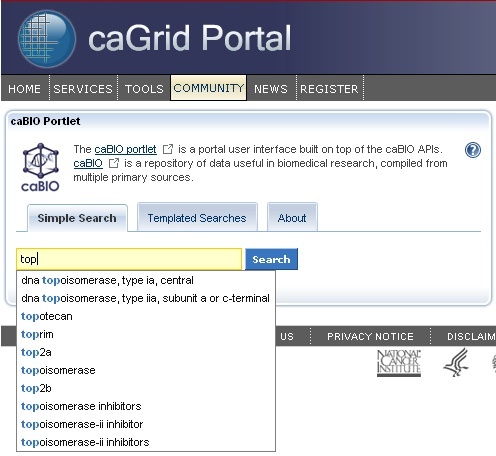
Selecting a Gene Search Term
The caBIO Portlet Simple Search tool creates an auto-populated list of caBIO terms that match the characters you have entered (1). Thus, you may select any of these suggested terms that match your disease concept of interest (for example, "TOP2A") to search for objects that match the term. If you would like to use a more general search term in order to retrieve objects that are associated with more than one disease term (for example, any cancer term associated with the ovary), you may use the "*" wild card character, which will replace zero or more characters (for example, a search for "TOP*").
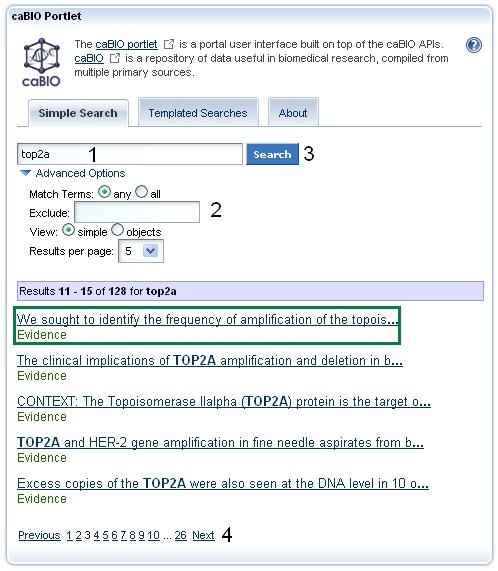
Simple Search Tool Retrieved Results
Once you have found a search term (1), click the Search button (2). If you would like, you may refine your search by clicking the Advanced Options (3). The Exclude field allows you to exclude objects that match your desired terms. You may select the format for retrieved results as lists of "simple" records, the default setting, or as objects. The Simple view returns at most three pieces of information from each retrieved object: the object name in blue text, miscellaneous data specific to the object's type in black text, and the type name in green text. You can view all of the object's attributes and data by clicking on the blue object name link. Alternatively, the Objects view will give a list of all attributes and metadata from retrieved objects.
Note
Due to space constraints and the nature of the Simple Search (that is, you must click through to a separate caBIO viewer to find the desired diseases and compounds), you will likely not find this view to be a useful option.
You may also increase the number of results per page from the default value of five to either ten or fifteen. Finally, if you enter multiple keywords, you may define whether to search for objects with attributes that contain any of the keywords, the default setting or that contain all of the keywords. This advanced option is only useful if you do not take advantage of the auto-populated list of matched search options.
Search Tip
It is strongly advised for you to select 15 returned results per page from the Advanced Options.
Simple Search queries retrieve lists of caBIO objects. The list is formatted such that, in general, three pieces of information are shown for each object - a blue link to all of the object's attributes and data, type-specific information (not present for these objects) in black text, and the object's type in green text. You do not have to first search the NCI Thesaurus to find an appropriate search term or code. Review the list of retrieved objects, clicking through to subsequent results pages, until you find an Evidence type object (green box).
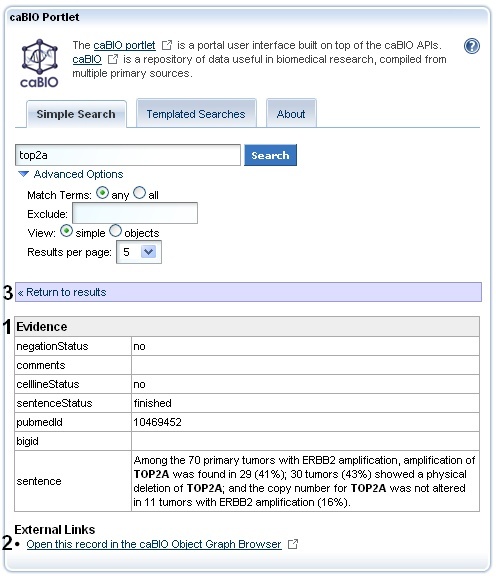
Once you click on the object's name, you may review the sentence, sentence status flag, cell line indicator, and negation indicator data for the object (1). In order to view the gene associated with this piece of evidence, you must use the caBIO Object Graph Browser.
Warning!
If you do not want to spend time navigating through the caBIO object model for candidate gene-disease or gene-compound/agent associations that were found to be false positives, unclear, or redundant to other data, you should first view the check that the sentenceStatus attribute is set to finished and the negationIndicator to no (1) before opening the caBIO Viewer (2).
To open the browser, click the Open this record in the caBIO Object Graph Browser link at the bottom of the page (2). A new window or tab in your web browser should open and display the Evidence type object. To return to the list of retrieved results, you may return to the Simple Search window or tab in your web browser and click the "Return to results" link (3).
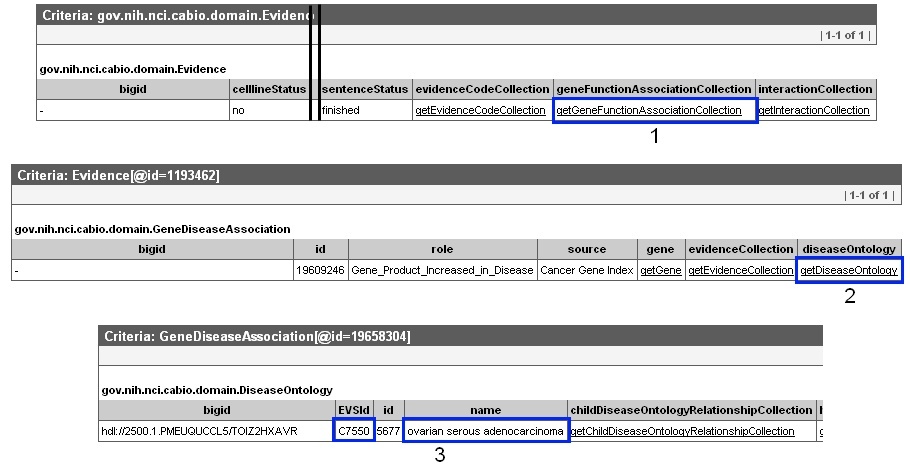
You must scroll to the right, represented as the black double line breaks in the Browser record, and click the getGeneFunctionAssocationCollection method link (1, blue box). You will then view links to access the Diseases or Genes (or even possibly both) that are associated with the gene and evidence, which will be grouped by the GeneDiseaseAssociation or GeneAgentAssociation type. To find the disease associated with a GeneDiseaseAssociation type object, click the getDiseaseOntology method link (2, blue box). You then can view the associated disease term name and EVS Identifier (3, blue box). To find the disease associated with a GeneAgentAssociation type object, click the getAgent method link, and view the associated compound term name and EVS Identifier (not shown).