|
Page History
...
Infrastructure for Algorithm Comparisons, Benchmarks, and Challenges, in Medical Imaging
| Panel | borderColor | dark red|
|---|---|---|
| ||
|
Author: Jayashree Kalpathy-Cramer
Introduction
Challenges are being increasingly viewed as a mechanism to foster advances in a number of domains including healthcare and medicine. The US Federal government, as part of the open government initiative has underscored the role of challenges https://www.whitehouse.gov/sites/default/files/docs/us_national_action_plan_6p.pdf as a way to "promote innovation through collaboration and (to) harness the ingenuity of the American Public." Large quantities of publicly available data and cultural changes in the openness of science have now made it possible to use these challenges and crowdsourcing efforts to propel the field forward.
...
In this article, we review a few historical imaging challenges. We then list the requirements we believe to be necessary (and nice to have) to support large-scale multimodal imaging challenges. We then review existing systems and develop a matrix of features and tools. Finally, we make some recommendations for developing Medical Imaging Challenge Infrastructure (MedICI), a system to support medical imaging challenges.
Review of historical challenges
...
Challenges have been popular in a number of scientific communities since the 1990s. In the text retrieval community, the Text REtrieval Conference (TREC) http://trec.nist.gov/ , co-sponsored by NIST is an early example of evaluation campaigns where participants work on a common task using data provided by the organizers and evaluated with a common set of metrics. ChaLearn http://www.chalearn.org/challenges.html has organized challenges in machine learning since 2013.
...
- A task is defined (the output). In our context, this could be segmentation of a lesion or organ, classification of an imaging study as being benign or malignant, prediction of survival, classification of a patients as being a responder or non-responder, pixel/voxel level classification of tissue or tumor grading.
- A set of images are provided (the input). These images are chosen to be of a sufficient size and diversity to reflect the challenges of the clinical problem. Data is typically spilt up into training and test datasets. The "truth" is made available to the participants for the training data but not the test data. This reduces the risk of overfitting the data and ensures the integrity of the results.
- An evaluation procedure is clearly defined; given the output of an algorithm on a the test images, one or more metrics are computed that measure the performance, usually a reference output is used in this process, but it could also be a visual evaluation of the results by human experts)
- Participants apply their algorithm to all data in the public test dataset provided. They can estimate their performance on the training test.
- Some challenges have an optional leaderboard phase where a subset of the test images is made available to the participants ahead of the final test. Participants can submit their results to the challenge system and have them evaluated or ranked but these are not considered the final standing.
- The reference standard or "ground truth" is defined using methodology clearly described to the participants but is not made publicly available in order to ensure that algorithm results are submitted to the organizers for publication rather than retained privately.
- Final valuation is carried out by the challenge organizers on the test set where the ground truth is sequestered from the participants.
Radiology and Pathology challenges for brain tumor imaging at MICCAI 2014
MICCAI 2014 held a day-long cluster of event in brain tumor computation including challenges for brain tumor classification and segmentation. The challenge consisted of radiology as well as pathology images. A majority of the images in the training data were from TCIA. Infrastructure support for the radiology portion of the challenges was provided by Bern University and the Virtual Skeleton Database system. The PAIS system support was provided by Stony Brook University for the pathology imaging.
...
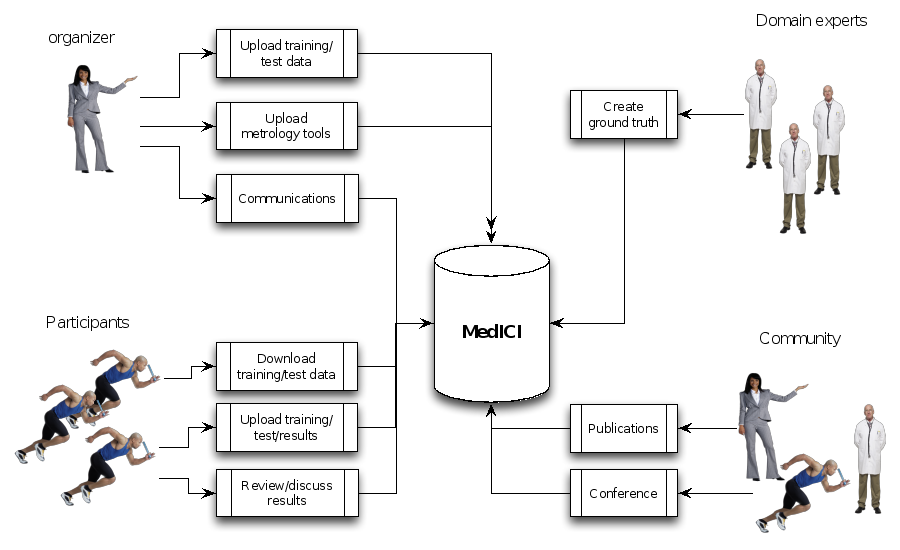
Figure 4 Challenge stakeholders and their tasks
Existing challenge infrastructure
A number of platforms exist for conducting challenges and crowdsourced efforts. Many of the popular platforms are commercial products, typically offering hosting and organizing services. Challenge organizers work with the company to set up the challenge. In some cases, the challenges are fairly trivial to set up and can be set up with the organizer without much support from the challenge platform company.
Commercial/hosted
We begin with a brief review a number of popular platforms used for challenges.
...
Challenge Post (http://challengepost.com/ ) has been used to organize hackathons, online challenges and other software collaborative activities. In person hackathons are free while the online challenges cost $1500/month (plus other optional chargesWhere does the section for commercial challenge management systems end?Could the suitability of each platform for imaging challenges be discussed?).
Open Source
Synapse is both an open source platform and a hosted solution for challenges and collaborative activities created by Sage bionetworks. It has been used for a number of challenges including the DREAM challenge. Synapse allows the sharing of code as well as data. However, the code typically is in R, Python and similar languages. Synapse also has a nice programmatic interface and methods to upload/download data, submit results, create annotations and provenance through R, Python, command line and Java. These options can be configured for the different challenges. Content in Synapse is referenced by unique Synapse IDs. The three basic types of Synapse objects include projects, folders and files. These can be accessed through the web interface or through programmatic APIs. Experience and support for running image analysis code within Synapse is limited.
...
| Kaggle/ | Synapse | HubZero (challenges/projects) | COMIC | VISCERAL | CodaLab |
Ease of setting up new challenge | 2/4 (if new metrics need to be used) | 2 | 2/5 | 2 | 3 | 1Explain the scoring system |
Cost (own server/hosting options) | $10-$25k/challenge | Free/hosted | Free/hosted | Free/hosted | Free/Azure costs | Free/hosted |
License | Commercial | OS | OS | OS | OS | OS |
Ease of extensibility | 5 | 4 | 4 | 2 | 3 | 2 |
Cloud support for algorithms | 4 | 3 | 3 | 4 | 1 | 3 |
Maturity | 1 | 1 | 1/5 | 3 | 4 | 3 |
Flexibility |
|
|
|
|
|
|
Number of users | 1 | 1 | 1/5 | 3 | 3 | 3 |
Types of challenges | 1 | 1 | 1 | 3 | 1 | 1 |
Native imaging support | No | No | No | Yes | Limited | No |
API to access data, code | 5 | 1 | 3 | 4 | 4 | 4 |
Components of challenge infrastructure
We describe below the various components of challenge infrastructure that would be necessary to host joint radiology/pathology challenges.
The web portal is the single point of entry for the participants. Historically, this would have information about the challenge, potentially host the data and provide a submission site for the user to upload results. The challenge organizer could also provide the results of the challenge at this page. Many challenges have wikis and announcement pages as well as forums. A good example of active discussion forums can be found at the Kaggle (https://www.kaggle.com/c/diabetic-retinopathy-detection/forums ). Most systems have backend systems (typically a relational database) for managing data and users. These allow registered used to access perhaps the training data and ground truth, the test data but not the ground truth. Challenge systems tailored for radiology and pathology also have specialized tools for handling these specialized data types and for creating and management of annotations and ground truth. Challenge systems also need modules for scoring and evaluation of the submissions. Finally, it is important to present the results back to the participants. Often these are presented in an ordered fashion with "winners at the top of the list.
Conclusions: Trade offs and Recommendations
Challenges have a very important role in moving science forward. In this document, we reviewed some of the more popular platforms to host challenges and compare some of the key aspects of these platforms. We believe that challenge infrastructure should be modular, flexible, extensible and user friendly. Requirements for this platform included support for radiology and pathology challenges. We were primarily seeking an open-source option. Although no single platform met all the requirements for our purposes, we were seeking solutions that could be extended easily and potentially had good interfaces that could be used to tie components together. We were envisioning potentially one solution for the more general aspects of challenge management (user and organizer management, data download, results upload, evaluation, results display), while adding other modules that are more specific for radiology and pathology imaging. A modular solution would allow us to switch out components as technologies mature or new technologies emerge.
...
Based on the time frame of this project and the current state of development of the platforms that we evaluated, we chose the CodaLab platform for the first iteration of MedICI as we believed that it offered the greatest compromise between current features, ease of use, flexibility and extensibility. However, we will re-evaluate this decision in 3 months and believe that we could very easily port any code we develop to other platforms.
For the radiology annotations and metadata management, we chose ePad while we chose caMicroscope for the pathology annotation and metadata management.
Reference
- Shi S, Pei J, Sadreyev RI, Kinch LN, Majumdar I, Tong J, Cheng H, Kim BH, Grishin NV. Analysis of CASP8 targets, predictions and assessment methods. Database : the journal of biological databases and curation. 2009;2009:bap003. doi: 10.1093/database/bap003. PubMed PMID: 20157476; PubMed Central PMCID: PMC2794793.
- Brownstein CA, Beggs AH, Homer N, Merriman B, Yu TW, Flannery KC, DeChene ET, Towne MC, Savage SK, Price EN, Holm IA, Luquette LJ, Lyon E, Majzoub J, Neupert P, McCallie D, Jr., Szolovits P, Willard HF, Mendelsohn NJ, Temme R, Finkel RS, Yum SW, Medne L, Sunyaev SR, Adzhubey I, Cassa CA, de Bakker PI, Duzkale H, Dworzynski P, Fairbrother W, Francioli L, Funke BH, Giovanni MA, Handsaker RE, Lage K, Lebo MS, Lek M, Leshchiner I, MacArthur DG, McLaughlin HM, Murray MF, Pers TH, Polak PP, Raychaudhuri S, Rehm HL, Soemedi R, Stitziel NO, Vestecka S, Supper J, Gugenmus C, Klocke B, Hahn A, Schubach M, Menzel M, Biskup S, Freisinger P, Deng M, Braun M, Perner S, Smith RJ, Andorf JL, Huang J, Ryckman K, Sheffield VC, Stone EM, Bair T, Black-Ziegelbein EA, Braun TA, Darbro B, DeLuca AP, Kolbe DL, Scheetz TE, Shearer AE, Sompallae R, Wang K, Bassuk AG, Edens E, Mathews K, Moore SA, Shchelochkov OA, Trapane P, Bossler A, Campbell CA, Heusel JW, Kwitek A, Maga T, Panzer K, Wassink T, Van Daele D, Azaiez H, Booth K, Meyer N, Segal MM, Williams MS, Tromp G, White P, Corsmeier D, Fitzgerald-Butt S, Herman G, Lamb-Thrush D, McBride KL, Newsom D, Pierson CR, Rakowsky AT, Maver A, Lovrecic L, Palandacic A, Peterlin B, Torkamani A, Wedell A, Huss M, Alexeyenko A, Lindvall JM, Magnusson M, Nilsson D, Stranneheim H, Taylan F, Gilissen C, Hoischen A, van Bon B, Yntema H, Nelen M, Zhang W, Sager J, Zhang L, Blair K, Kural D, Cariaso M, Lennon GG, Javed A, Agrawal S, Ng PC, Sandhu KS, Krishna S, Veeramachaneni V, Isakov O, Halperin E, Friedman E, Shomron N, Glusman G, Roach JC, Caballero J, Cox HC, Mauldin D, Ament SA, Rowen L, Richards DR, San Lucas FA, Gonzalez-Garay ML, Caskey CT, Bai Y, Huang Y, Fang F, Zhang Y, Wang Z, Barrera J, Garcia-Lobo JM, Gonzalez-Lamuno D, Llorca J, Rodriguez MC, Varela I, Reese MG, De La Vega FM, Kiruluta E, Cargill M, Hart RK, Sorenson JM, Lyon GJ, Stevenson DA, Bray BE, Moore BM, Eilbeck K, Yandell M, Zhao H, Hou L, Chen X, Yan X, Chen M, Li C, Yang C, Gunel M, Li P, Kong Y, Alexander AC, Albertyn ZI, Boycott KM, Bulman DE, Gordon PM, Innes AM, Knoppers BM, Majewski J, Marshall CR, Parboosingh JS, Sawyer SL, Samuels ME, Schwartzentruber J, Kohane IS, Margulies DM. An international effort towards developing standards for best practices in analysis, interpretation and reporting of clinical genome sequencing results in the CLARITY Challenge. Genome biology. 2014;15(3):R53. doi: 10.1186/gb-2014-15-3-r53. PubMed PMID: 24667040; PubMed Central PMCID: PMC4073084.
- Omberg L, Ellrott K, Yuan Y, Kandoth C, Wong C, Kellen MR, Friend SH, Stuart J, Liang H, Margolin AA. Enabling transparent and collaborative computational analysis of 12 tumor types within The Cancer Genome Atlas. Nature genetics. 2013;45(10):1121-6. doi: 10.1038/ng.2761. PubMed PMID: 24071850; PubMed Central PMCID: PMC3950337.
- Abdallah K, Hugh-Jones C, Norman T, Friend S, Stolovitzky G. The Prostate Cancer DREAM Challenge: A Community-Wide Effort to Use Open Clinical Trial Data for the Quantitative Prediction of Outcomes in Metastatic Prostate Cancer. The oncologist. 2015. doi: 10.1634/theoncologist.2015-0054. PubMed PMID: 25777346.
- Jarchum I, Jones S. DREAMing of benchmarks. Nat Biotechnol. 2015;33(1):49-50. doi: 10.1038/nbt.3115. PubMed PMID: 25574639.