|
Page History
...
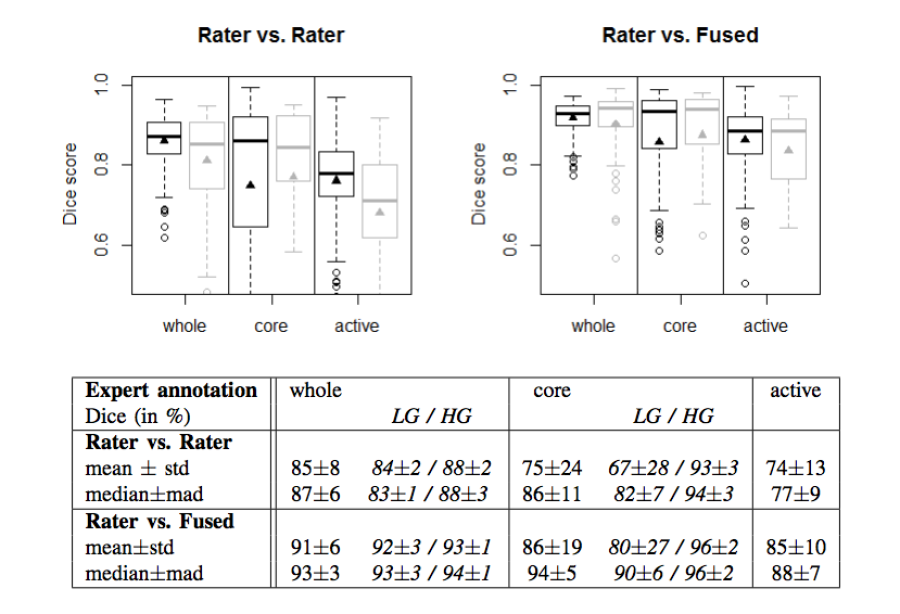
Figure 2. Dice coefficients of inter-rater agreement and of rater vs. fused label maps
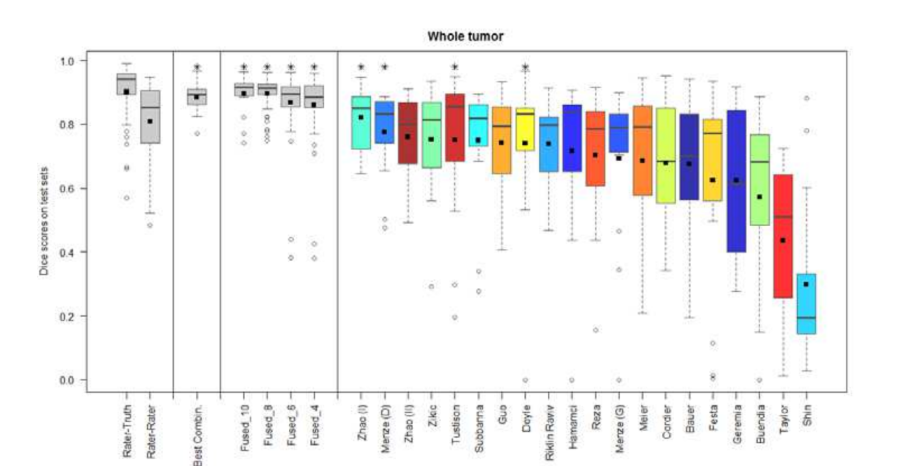
Figure 3. Dice coefficients of individual algorithms and fused results indicating improvement with label fusion
...
The HubZero is an open source platform developed for scientific collaboration. It has been used heavily in a number of communities including nanoscience, earthquake engineering, molecular diagnostics and others. A version focused on cancer informatics can is hosted at nciphub.org. nciphub shares a lot of features with the Synapse platform. It allows user management and role-based access. Users can create groups that share common interest and collaborate within these groups. Files can be shared within projects. Other features include wiki, calendars, creating and sharing resources such as presentations, multimedia and even tools. Most common tools found on the various hubs are those based on simulations. Although nciphub has limited native support of medical imaging, libraries to handle medical images can be configured to work in the hub. Members of the Quantitative Imaging Network (QIN) are exploring the use of nciphub for challenges, especially for the communication and data sharing.
CodaLab is an open-source project that originated at Microsoft Research that was expressly created for hosting challenges and supporting reproducible research. The OuterCurve Foundation currently maintains it. Challenge organizers can easily set up challenges by creating a competition bundle that consists of data as well as evaluate tools. As part of the configuration files, the number of phases and duration (e.g. training, leaderboard, test) can be set up by the organizer. The evaluation program can be written in any language. Participants can upload results and get immediate feedback. The currently available version of CodaLab comes with scoring algorithms for image segmentation evaluation Organizers can extend the presentation of results to allow drilling down into the results with tables and charts. CodaLab currently uses the Azure platform although, in theory, it should be possible to deploy on other servers without a great deal of effort. CodaLab is also developing support for worksheets. These are resources to support reproducible research and for collaboration. Using these, researchers have compared a number of open source NLP tools on different public datasets. As this technology continues to be developed, researchers will be able to quickly compare the performance of different algorithms on a range of datasets in the "cloud" by leveraging Azure technology.
...
Based on the time frame of this project and the current state of development of the platforms that we evaluated, we chose the CodaLab platform for the first iteration of MedICI as we believed that it offered the greatest compromise between current features, ease of use, flexibility and extensibility. However, we will re-evaluate this decision in 3 months and believe that we could very easily port any code we develop to other platforms.
For the radiology annotations and metadata management, we chose ePad while we chose caMicroscope for the pathology annotation and metadata management.
...