|
Page History
Infrastructure for Algorithm Comparisons, Benchmarks, and Challenges in Medical Imaging
AuthorAuthors: Jayashree Kalpathy-Cramer and Karl Helmer
...
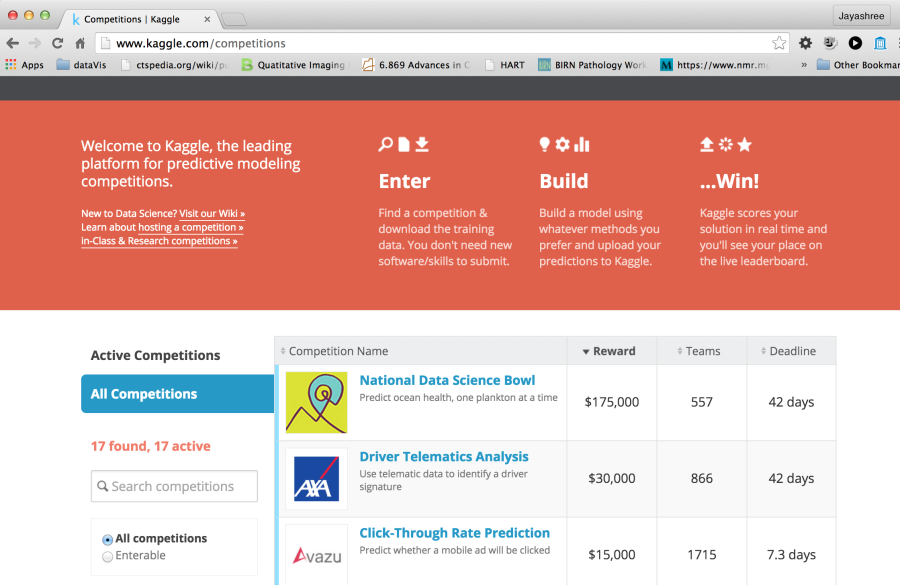
Figure 5. Portal for Kaggle, a leading website for challenges for data scientists
| Multiexcerpt include | ||||||
|---|---|---|---|---|---|---|
|
...
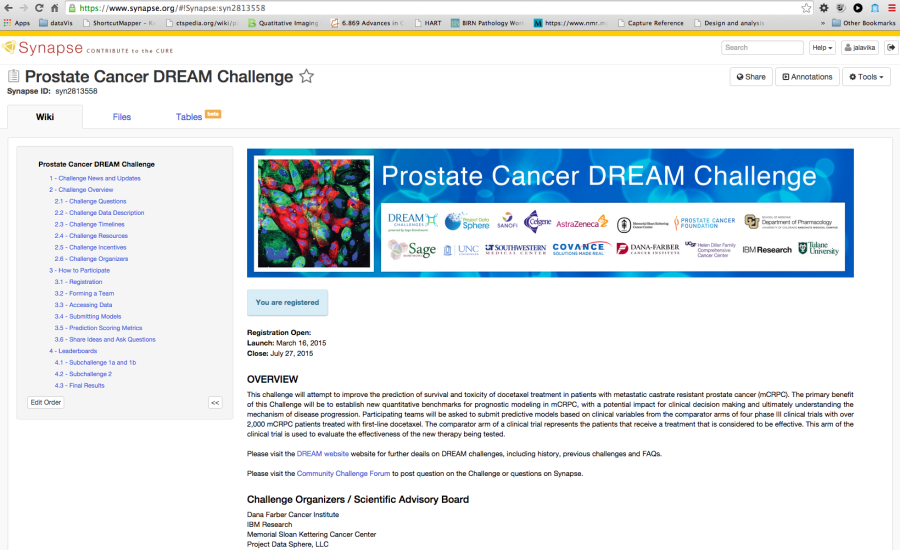
Figure 7. Example Challenge hosted in Synapse
| Multiexcerpt include | ||||||
|---|---|---|---|---|---|---|
|
The main steps to create a new challenge
| Multiexcerpt include | ||||||
|---|---|---|---|---|---|---|
|
- Create a project
Multiexcerpt include nopanel true MultiExcerptName ExitDisclaimer PageWithExcerpt wikicontent:Exit Disclaimer to Include - Add pages
Multiexcerpt include nopanel true MultiExcerptName ExitDisclaimer PageWithExcerpt wikicontent:Exit Disclaimer to Include - Making uploaded files available for download
- Allowing others to register for your project
Multiexcerpt include nopanel true MultiExcerptName ExitDisclaimer PageWithExcerpt wikicontent:Exit Disclaimer to Include - Allowing others to register for your project
Multiexcerpt include nopanel true MultiExcerptName ExitDisclaimer PageWithExcerpt wikicontent:Exit Disclaimer to Include - Make your Make your project appear in the projects overview
Multiexcerpt include nopanel true MultiExcerptName ExitDisclaimer PageWithExcerpt wikicontent:Exit Disclaimer to Include - Allow file uploads
- Including content from files on a page
- Allow others to edit the project
- Changing colors and other styling
- Project data folder
Multiexcerpt include nopanel true MultiExcerptName ExitDisclaimer PageWithExcerpt wikicontent:Exit Disclaimer to Include - Including content from files on a page
Multiexcerpt include nopanel true MultiExcerptName ExitDisclaimer PageWithExcerpt wikicontent:Exit Disclaimer to Include - Allow others to edit the project
Multiexcerpt include nopanel true MultiExcerptName ExitDisclaimer PageWithExcerpt wikicontent:Exit Disclaimer to Include - Changing colors and other styling
Multiexcerpt include nopanel true MultiExcerptName ExitDisclaimer PageWithExcerpt wikicontent:Exit Disclaimer to Include - Project data folder
Multiexcerpt include nopanel true MultiExcerptName ExitDisclaimer PageWithExcerpt wikicontent:Exit Disclaimer to Include - Page permissions
Page permissionsMultiexcerpt include nopanel true MultiExcerptName ExitDisclaimer PageWithExcerpt wikicontent:Exit Disclaimer to Include
However, at this time, there is limited support for automatic evaluation of submitted results, results presentation, native support for medical images although many of these features are planned.
The HubZero
| Multiexcerpt include | ||||||
|---|---|---|---|---|---|---|
|
| Multiexcerpt include | ||||||
|---|---|---|---|---|---|---|
|
| Multiexcerpt include | ||||||
|---|---|---|---|---|---|---|
|
...
The MIDAS platform has been used to host a couple of imaging challenges. A special
| Multiexcerpt include | ||||||
|---|---|---|---|---|---|---|
|
...
The web portal is the single point of entry for the participants. Historically, this would have information about the challenge, potentially host the data and provide a submission site for the user to upload results. The challenge organizer could also provide the results of the challenge at this page. Many challenges have wikis and announcement pages as well as forums. A good example of active discussion forums can be found at the Kaggle
| Multiexcerpt include | ||||||
|---|---|---|---|---|---|---|
|
...