|
Page History
...
Our next step is to validate all the files, ; which we will do in two passes: the first pass, validating only the TXT files (but not the IDF and SDRF), and the second pass, validating all the files (IDF, so by checking off every single file in the list (IDF, SDRF, and TXT). For the first pass, check off all the TXT files in the list, then click then clicking the 'Validate' button below.
...
The 'Manage Data' tab now refreshes with the status of the array data files showing as 'In Queue'.
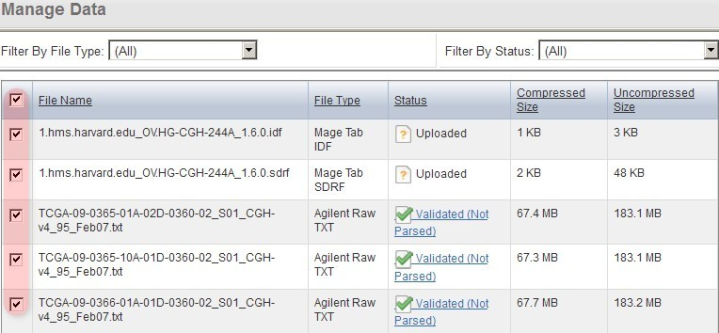
You'll know when the first pass validation is successful when the status for TXT of the files shows as 'Validated' or 'Validated, Not Parsed'. (NOTE: The 'Not Parsed' status doesn't indicate any problem with your array data; caArray still has not would only show in versions of caArray prior to v2.4.0 which had not yet implemented a parser for the Agilent TXT format and is were thus unable to parse these files. The These files can still be imported into your experiment without being parsed.)
...
Once the data finishes validating, the 'Manage Data' tab will appear with the status of the array data files showing as 'Validated (Not Parsed)'.
For the second pass, check off every single file on the 'Manage Data' page (IDF, SDRF, and TXT), then again click the 'Validate' button below.
To finish verifying the uploaded data, check off all the files under the 'Manage Data' tab (IDF, SDRF, and array data files), then click the 'Validate' button again.
...