|
Page History
...
| Panel | ||||
|---|---|---|---|---|
| ||||
|
| Anchor | ||||
|---|---|---|---|---|
|
Introduction to CTIIP
Most cancer diagnoses are made based on images. You have to see a tumor, or compare images of it over time, to determine its level of threat. Ultrasounds, MRIs, and X-rays are all common types of images that radiologists use to collect information about a patient and perhaps cause a doctor to recommend a biopsy. Once that section of the tumor is under the microscope, pathologists learn more about it. Radiologists and pathologists represent different scientific disciplines. To gather even more information, a doctor may order a genetic panel. If that panel shows that the patient has a genetic anomaly, the doctor or a geneticist may search for clinical trials that match it, or turn to therapies that researchers have already proven effective for this combination of tumor and genetic anomaly through recent advances in precision medicine.
...
Within the three research domains that CTIIP intends to make available for integrative queries, only one, clinical imaging, has made some progress in terms of establishing a framework and standards for informatics solutions. Those standards include Annotation and Image Markup (AIM), which allows researchers to standardize annotations and markup for radiology images, and Digital Imaging and Communications in Medicine (DICOM), which is a standard for handling, storing, printing, and transmitting information in medical imaging. For pre-clinical imaging and digital pathology, there are no such standards that allow for the seamless viewing, integration, and analysis of disparate data sets to produce integrated views of the data, quantitative analysis, data integration, and research or clinical decision support systems. The micro-Annotation and Image Markup (µAIM) model is currently in development to serve the unique needs of the pre-clinical domain.
...
| Domain | Data Set | Applicable Standard | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Molecular and Clinical ImagingData | The Cancer Genome Atlas (TCGA) molecular and clinical and molecular data | N/A | |||||||||
| Clinical Imaging | The Cancer Imaging Archive (TCIA) in vivo imaging data | DICOM | |||||||||
| Pre-Clinical | Small animal models | N/A A standard exists but has not been adopted (ask Ulli) | Supplement 187: Preclinical Small Animal Imaging Acquisition Context
| ||||||||
| Digital Digital Pathology | caMicroscope | DICOM is applicable but has not yet been adopted(ask Ulli). | |||||||||
| All | Annotations and markup on images | µAIM is in development. |
Digital Pathology and Integrated Query System
One of the goals of this sub-project is to create a digital pathology image server that can accept whole slide images from multiple vendors and display them despite the proprietary formats they were created in. They This is accomplished by integrating the OpenSlide
| Multiexcerpt include | ||||||
|---|---|---|---|---|---|---|
|
...
The team is using OpenSlide, a vendor-neutral C library, to extend the software of caMicroscope, a digital pathology server. The extended software will support some of the common formats adopted by whole slide vendors as well as basic image analysis algorithms. With the incorporation of common whole slide formats, caMicroscope will be able to read whole slides without recoding, which often introduces additional compression artifacts, and provide a logical bridge from proprietary pathology formats to DICOM standards.
Image markups and annotations also require standards so that they can be read by different imaging disciplines along with the rest of the image data. Support for the micro-Annotation and Image Markup ( μ-AIM ) model will be added to caMicroscope so that researchers can include image annotation and markup features in digital pathology data.
...
Data federation, a process whereby data is collected from different databases without ever copying or transferring the original data, is part of the new infrastructure. The software used to accomplish this data federation is Bindaas. Bindaas is middleware that is also used to build the backend infrastructure of caMicroscope. The team is extending Bindaas with a data federation capability that makes it possible to query data from TCIA and TCGA. Bindaas is a middleware used to develop web services that allow data providers to share data, stored in databases, using a popular standard for developing web services called Representational State Transfer. Developers can use the REST interface with most modern languages to rapidly create and deploy applications that can consume data contained in the underlying database. Bindaas enables resource providers to rapidly generate APIs with only an understanding of the underlying data model. It is able to do so because it uses a declarative programming model that allows data providers to create REST APIs without having to write a single line of code.
The Integrated Query System will access multiple data types in a federated fashion, meaning that the original data will reside in independent systems. The Integrated Query System will provide an interface scientists can use to select the data types they want to combine, or "mash up," based on their own research questions.
...
Challenges are often conducted in conjunction with scientific conferences. The following Pilot Challenges, supported by the CTIIP project and described in the following table, were part of the Computational Brain Tumor Cluster of Event (CBTC) 2015 which was held on October 9, 2015 in Munich, Germany, in conjunction with MICCAI 2015.
| MICCAI 2015 Challenges | Sample Image | Description | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Combined Radiology and Pathology Classification
| The datasets for this challenge are Radiology and Pathology images obtained from the same patients. Each case corresponds to a single patient. There is one Radiology image and one whole slide tissue image for each case. The training set contains a total of 32 cases: 16 cases that are classified by pathologists as Oligodendroglioma and 16 cases classified as Astrocytoma. The test set will have 20 cases. Please note that the number of cases in the test set may not be equally partitioned between the two sub-types. The whole slide tissue images are stored in Aperio SVS format. There are open source tools and libraries that can read these images: OpenSlide
| |||||||||||||||||
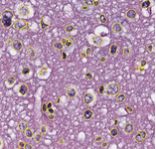
Segmentation of Nuclei in Pathology Images
| The goal of this challenge is to evaluate the performance of algorithms for detection and segmentation of nuclear material in a tissue image. Participants are asked to detect and segment all the nuclei in a given set of image tiles extracted from whole slide tissue images. The algorithm results will be compared with consensus pathologist-segmented sub-regions. Winners will be ranked based on their nuclei segmentation best matching the reference standards. The reference standard for the challenge will be pathologist-generated nuclear segmentation on select regions of TCGA Glioma whole slide images. |
...