Introduction to CTIIP
Most cancer diagnoses are made based on images. You have to see a tumor, or compare images of it over time, to determine its level of threat. Ultrasounds, MRIs, and X-rays are all common types of images that radiologists use to collect information about a patient and perhaps cause a doctor to recommend a biopsy. Once that section of the tumor is under the microscope, pathologists learn more about it. Radiologists and pathologists represent different scientific disciplines. To gather even more information, a doctor may order a genetic panel. If that panel shows that the patient has a genetic anomaly, the doctor or a geneticist may search for clinical trials that match it, or turn to therapies that researchers have already proven effective for this combination of tumor and genetic anomaly through recent advances in precision medicine.
Another way we learn about cancer in humans is through small animal research. Images from small animals allow detailed study of biological processes, disease progression, and response to therapy, with the potential to provide a natural bridge to human disease. Due to differences in how data is collected and stored about animals and humans, however, the bridge is man-made.
Each of these diagnostic images are at a different scale, from a different scientific discipline. A large-scale image like an X-ray may be almost life-size. Slices of tumors are smaller still and you must put them on a slide under a microscope to see them. Not surprisingly, each of these image types require specialized knowledge to create, handle, and interpret them. While complementary, each specialist comes from a different scientific discipline.
One promise of big data is that data mashups can integrate two or more data sets in a single interface so that doctors, pathologists, radiologists, and laboratory technicians can make connections that improve outcomes for patients. Such mashups require and await technical solutions in the areas of data standards and software development. A significant start to all of these technical solutions are the sub-projects of the National Cancer Institute Clinical and Translational Imaging Informatics Project (NCI CTIIP).
CTIIP Sub-Projects
As discussed so far, cancer research is needed across disciplines. To serve this need, the National Cancer Institute Clinical and Translational Imaging Informatics Project (NCI CTIIP) team plans to meet it by creating a data mashup interface, along with other software and standards, that accesses The Cancer Genome Atlas (TCGA) clinical and molecular data, The Cancer Imaging Archive (TCIA) in-vivo imaging data, caMicroscope pathology data, a pilot data set of animal model data, and relevant imaging annotation and markup data.
The common informatics infrastructure that will result from this project will provide researchers with analysis tools they can use to directly mine data from multiple high-volume information repositories, creating a foundation for research and decision support systems to better diagnose and treat patients with cancer.
CTIIP is composed of the following sub-projects. Each project is discussed on this page.
| Sub-Project Name | Description |
|---|---|
| Digital Pathology | Addresses the accessibility of digital pathology data through the integration of OpenSlide, improves tools for annotation and markup of pathology images through the development of microAIM (μ-AIM), and integrates analysis tools with caMicroscope. These developments increase the interoperability in each of the targeted research domains: clinical imaging, pre-clinical imaging, and digital pathology imaging. |
| Integrated Query System | Provides a data mashup interface that accesses TCGA clinical and molecular data, TCIA in-vivo imaging data, caMicroscope pathology data, relevant imaging annotation and markup data, and a pilot data set of animal model data. |
| DICOM Standards for Small Animal Imaging; Use of Informatics for Co-clinical Trials | Addresses the need for standards in pre-clinical imaging and applies the informatics tools created by the Digital Pathology and Integrated Query System sub-projects to co-clinical trials. |
| Pilot Challenges | Pilot challenges are a tool to find suitable image analysis algorithms. The pilot challenges would use limited data sets for proof-of-concept, and test the informatics infrastructure needed for more rigorous “Grand Challenges” that could later be scaled up and supported by extramural initiatives. |
The Importance of Data Standards
NCI CBIIT has worked extensively for several years in the area of data standards for both clinical research and healthcare, working with the community and Standards Development Organizations (SDOs), such as the Clinical Data Interchange Standards Consortium (CDISC), Health Level 7 (HL7) and the International Organization for Standardization (ISO). From that work, Enterprise Vocabulary Services (EVS) and Cancer Data Standards Registry and Repository (caDSR) are harmonized with the Biomedical Research Integrated Domain Group (BRIDG), Study Data Tabulation Model (SDTM), and Health Level Seven® Reference Information Model HL7 RIM models. Standardized Case Report Forms (CRFs), including those for imaging, have also been created. The CBIIT project work provides the bioinformatics foundation for semantic interoperability in digital pathology and co-clinical trials integrated with clinical and patient demographic data and data contained in TCIA and TCGA.
The common infrastructure that will result from CTIIP and its sub-projects depends on data interoperability, which is greatly aided by adherence to data standards. While image data standards exist to support communicating image data in a common way, the data standards that do exist for image data are inconsistently adopted. One reason for the lack of uniform adoption is that vendors of image management tools required for the analysis of imaging data have created these tools so that they only accept proprietary data formats. Researchers then make sure their data can be interpreted by these tools. The result is that data produced on different systems cannot be analyzed by the same mechanisms.
Another challenge for CTIIP with its goal of integrating data from complimentary domains is the lack of a defined standard for co-clinical and digital pathology data. Without a data standard for these domains, it is very difficult to share and leverage such data across studies and institutions. As part of the CTIIP project, the team has extended the DICOM model to co-clinical and small animal imaging. The long-term goal is to generate DICOM-compliant images for small animal research.
Within the three research domains that CTIIP intends to make available for integrative queries, only one, clinical imaging, has made some progress in terms of establishing a framework and standards for informatics solutions. Those standards include Annotation and Image Markup (AIM), which allows researchers to standardize annotations and markup for radiology images, and Digital Imaging and Communications in Medicine (DICOM), which is a standard for handling, storing, printing, and transmitting information in medical imaging. For pre-clinical imaging and digital pathology, there are no such standards that allow for the seamless viewing, integration, and analysis of disparate data sets to produce integrated views of the data, quantitative analysis, data integration, and research or clinical decision support systems. The micro-Annotation and Image Markup (µAIM) model is currently in development to serve the unique needs of the pre-clinical domain.
The following table presents the data that the CTIIP team is integrating through various means. This integration relies on the expansion of software features and on the application of data standards, as described in subsequent sections of this document.
| Domain | Data Set | Applicable Standard |
|---|---|---|
| Molecular and Clinical Data | The Cancer Genome Atlas (TCGA) molecular and clinical data | N/A |
| Clinical Imaging | The Cancer Imaging Archive (TCIA) in vivo imaging data | DICOM |
| Pre-Clinical | Small animal models | Supplement 187: Preclinical Small Animal Imaging Acquisition Context of the DICOM standard exists but has not yet been adopted. |
| Digital Pathology | caMicroscope | DICOM is applicable but has not yet been adopted. |
| All | Annotations and markup on images | µAIM is in development. |
Digital Pathology and Integrated Query System
One of the goals of this sub-project is to create a digital pathology image server that can accept whole slide images from multiple vendors and display them despite the proprietary formats they were created in. This is accomplished by integrating the OpenSlide libraries with caMicroscope.
Using this server, which is an extended version of caMicroscope, researchers can select data from different imaging data sets and use them in image algorithms. The first data sets that are being integrated on this image server are TCGA and TCIA.
TCGA finalized tissue collection with matched tumor and normal tissues from 11,000 patients, allowing for the comprehensive characterization of 33 cancer types and subtypes, including 10 rare cancers, and has provided this information to the research community. TCIA and the underlying National Biomedical Image Archive (NBIA) manage well-curated, publicly-available collections of medical image data. The linkages between TCGA and TCIA are valuable to researchers who want to study diagnostic images associated with the tissue samples sequenced by TCGA. TCIA currently supports over 40 active research groups including researchers who are exploiting these linkages.
Although TCGA and TCIA comprise a rich, complementary, multi-discipline data set, they are in an infrastructure that provides limited ability to query the data. Researchers want to query multiple databases at the same time to identify cases based on all available data types.
To address these limitations, the CTIIP team is developing an Integrated Query System to make it easier to analyze data from different research disciplines represented by TCGA, TCIA, and co-clinical/small animal model data.
Digital Pathology
Digital pathology, unlike its more mature radiographic counterpart, has yet to standardize on a single storage and transport media. In addition, each pathology-imaging vendor produces its own image management system, making image analysis systems proprietary and not standardized. The result is that images produced on different systems cannot be viewed and analyzed via the same mechanisms. Not only does this lack of standards and the dominance of proprietary formats impact digital pathology, but it prevents digital pathology data from integrating with data from other disciplines.
The team is using OpenSlide, a vendor-neutral C library, to extend the software of caMicroscope, a digital pathology server. The extended software will support some of the common formats adopted by whole slide vendors as well as basic image analysis algorithms. With the incorporation of common whole slide formats, caMicroscope will be able to read whole slides without recoding, which often introduces additional compression artifacts.
Image markups and annotations also require standards so that they can be read by different imaging disciplines along with the rest of the image data. Support for the μ-AIM model will be added to caMicroscope so that researchers can include image annotation and markup features in digital pathology data.
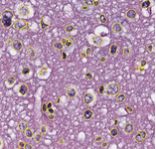
caMicroscope Slide with Markup
With caMicroscope's support for basic image analysis algorithms, researchers can use this tool to enable analytic and decision support using digital pathology images.
Integrated Query System
The purpose of the integrative query component of CTIIP is to support data mashups between images, image-derived information, and clinical, pre-clinical, and genomic data. Co-clinical data and clinical data such as patient information and outcome will also be accessible through the Integrated Query System.
To make data accessible and comparable, it must first be collected in a structured fashion. For example, TCGA relies on Common Data Elements, which are the standard elements that structure TCGA data. Second, data comparisons require common data vocabularies. For example, when a tumor is described in a human or an animal, one of a discrete number of approved vocabulary options must be used to describe the tumor.
Data federation, a process whereby data is collected from different databases without ever copying or transferring the original data, is part of the new infrastructure. The software used to accomplish this data federation is Bindaas. Bindaas is a middleware used to develop web services that allow data providers to share data, stored in databases, using a popular standard for developing web services called Representational State Transfer. Developers can use the REST interface with most modern languages to rapidly create and deploy applications that can consume data contained in the underlying database. Bindaas enables resource providers to rapidly generate APIs with only an understanding of the underlying data model. It is able to do so because it uses a declarative programming model that allows data providers to create REST APIs without having to write a single line of code.
The Integrated Query System will access multiple data types in a federated fashion, meaning that the original data will reside in independent systems. The Integrated Query System will provide an interface scientists can use to select the data types they want to combine, or "mash up," based on their own research questions.
The following table presents the data types and their sources that the Integrated Query System will make available.
| Data Types in the Integrated Query System | Data Source |
|---|---|
| Genomic | Google Genomics Cloud |
| Clinical | Downloaded from TCGA and stored in a customized database at Emory University |
| Preclinical | Customized database at Emory University |
| Radiology Images (Human and Animal) | TCIA |
| Radiology Image Annotation and Markup | AIM Data Service (AIME) |
| Pathology Images (Human and Animal) | caMicroscope |
| Pathology Image Annotation and Markup | uAIM Data Service (uAIME) |
The Integrated Query System, with its support for whole slides and data mashups of federated data, will act as a foundation for a broader set of novel community research projects.
DICOM Working Group 30
Since its first publication in 1993, DICOM has revolutionized the practice of radiology, allowing the replacement of X-ray film with a fully digital workflow. Each year, the standard is updated with formats for medical images that can be exchanged with the data and quality necessary for clinical use. (Source: http://dicom.nema.org/Dicom/about-DICOM.html)
As part of the Small Animal/Co-clinical Improved DICOM Compliance and Data Integration sub-project of CTIIP, the NCI supported the development of a DICOM supplement for small animal imaging. The group of people contributing to it, Working Group 30, completed Supplement 187: Preclinical Small Animal Imaging Acquisition Context , in 2015.
Supplement 187 Data Elements
Information about how a small animal image was acquired is relevant to the interpretation of the image and must be stored with it. While DICOM defines terminology applicable to other types of images, it does not include data elements associated with small animal image acquisition. The new Supplement 187, developed as part of the CTIIP project in 2015, defines terminology that is unique to small animal imaging. It includes the following templates that include terminology relevant to image acquisition.
- Preclinical Small Animal Image Acquisition Context
- Language of Content Item and Descendants
- Observation Context
- Biosafety Conditions
- Animal Housing
- Animal Feeding
- Heating Conditions
- Circadian Effects
- Physiological Monitoring Performed During Procedure
- Anesthesia
- Medications and Mixture Medications
- Medication, Substance, Environmental Exposure
Consult Supplement 187: Preclinical Small Animal Imaging Acquisition Context for details about each of these templates.
Pilot Challenges
Challenges are being increasingly viewed as a mechanism to foster advances in a number of fields, including healthcare and medicine. Large quantities of publicly available data, such as that in TCIA, and cultural changes in the openness of science have now made it possible to use these challenges, as well as crowdsourcing (enlisting the services of people via the Internet), to propel the field forward.
Some of the key advantages of challenges over conventional methods include 1) scientific rigor (sequestering the test data), 2) comparing methods on the same datasets with the same, agreed-upon metrics, 3) allowing computer scientists without access to medical data to test their methods on large clinical datasets, 4) making resources available, such as source code, and 5) bringing together diverse communities (that may traditionally not work together) of imaging and computer scientists, machine learning algorithm developers, software developers, clinicians, and biologists.
As explained in the Challenge Management System Evaluation Report, challenge hosts and participants cannot do it alone. The computing resourcing needed to process these large datasets may be beyond what is available to individual participants. For the organizers, creating an infrastructure that is secure, robust, and scalable can require resources that are beyond the reach of many researchers. Additionally, imaging formats for pathology images can be proprietary and interoperability between formats can require additional software development efforts.
Over the last few years, Grand Challenges have become popular in several imaging-based research communities. A Grand Challenge is designed to validate and compare imaging analysis algorithms. The algorithms are applied to a single dataset and the results for each algorithm are compared against a previously-determined ground-truth dataset.
The Pilot Challenges sub-project of CTIIP will sponsor complementary Pilot Challenge projects. As opposed to a more rigorous Grand Challenge, each Pilot Challenge will involve data sets of reduced size and demonstrate the infrastructure as capable of running Grand Challenges in the future.
Challenges are often conducted in conjunction with scientific conferences. The following Pilot Challenges, supported by the CTIIP project and described in the following table, were part of the Computational Brain Tumor Cluster of Event (CBTC) 2015 which was held on October 9, 2015 in Munich, Germany, in conjunction with MICCAI 2015.
| MICCAI 2015 Challenges | Sample Image | Description |
|---|---|---|
Combined Radiology and Pathology Classification
| The datasets for this challenge are Radiology and Pathology images obtained from the same patients. Each case corresponds to a single patient. There is one Radiology image and one whole slide tissue image for each case. The training set contains a total of 32 cases: 16 cases that are classified by pathologists as Oligodendroglioma and 16 cases classified as Astrocytoma. The test set will have 20 cases. Please note that the number of cases in the test set may not be equally partitioned between the two sub-types. The whole slide tissue images are stored in Aperio SVS format. There are open source tools and libraries that can read these images: OpenSlide and Bio-Formats . | |
Segmentation of Nuclei in Pathology Images
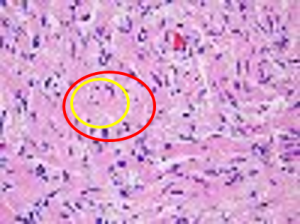
| The goal of this challenge is to evaluate the performance of algorithms for detection and segmentation of nuclear material in a tissue image. Participants are asked to detect and segment all the nuclei in a given set of image tiles extracted from whole slide tissue images. The algorithm results will be compared with consensus pathologist-segmented sub-regions. Winners will be ranked based on their nuclei segmentation best matching the reference standards. The reference standard for the challenge will be pathologist-generated nuclear segmentation on select regions of TCGA Glioma whole slide images. |
A team from Massachusetts General Hospital will guide the Pilot Challenges. They are using Medical Imaging Challenge Infrastructure (MedICI), a medical imaging challenge platform, to support the challenges. MedICI, in turn, uses the CodaLab framework, an open-source challenge platform developed by Microsoft Research and others in the medical imaging and machine learning communities. Because CodaLab does not have built-in imaging handling, display or annotation capabilities, the team is building on two application packages, ePad and caMicroscope, to provide those features. For example, once participants upload their results, they can see them in ePad.
Challenge participants receive test and training data by creating shared lists in TCIA, then pulling those into CodaLab.