|
Page History
If you need to move a large volume of data, you can do so using a Globus endpoint. DME uses Globus for high performance transfer of large files. Endpoints represent locations on a file transfer service server, such as Biowulf or gridftp. In DME CLU or GUI, you can choose to use Globus for file transfer. A prerequisite is a guest collection.
For narrated slides demonstrating these instructions, refer to prep-Globus-slides.pptx.
To prepare a Globus endpoint:
If you do not already have a shareable Globus endpoint, contact NCIDataVault@mail.nih.gov and request one.
HTML Comment hidden true When people send messages to the NCI Data Vault inbox, requesting an endpoint, Udit usually asks them various questions. I asked about including those questions here, but Sunita said most of them have never seen this page. People usually see this page after the DME team has talked with them.
- Log into Globus at the following site:
http://www.globus.orgMultiexcerpt include MultiExcerptName ExitDisclaimer nopanel true PageWithExcerpt wikicontent:Exit Disclaimer to Include
Use your NIH user account credentials as described in the following page:
https://docs.globus.org/how-to/get-started/Multiexcerpt include MultiExcerptName ExitDisclaimer nopanel true PageWithExcerpt wikicontent:Exit Disclaimer to Include If you do not already have a guest collection in Globus, create one:
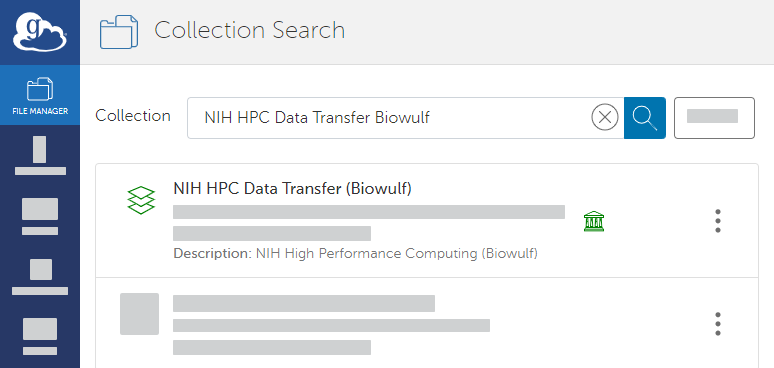
Panel borderColor black borderStyle solid Expand title Instructions to Create a Guest Collection - In the Globus File Manager, search for NIH HPC Data Transfer (Biowulf). This is a public endpoint.
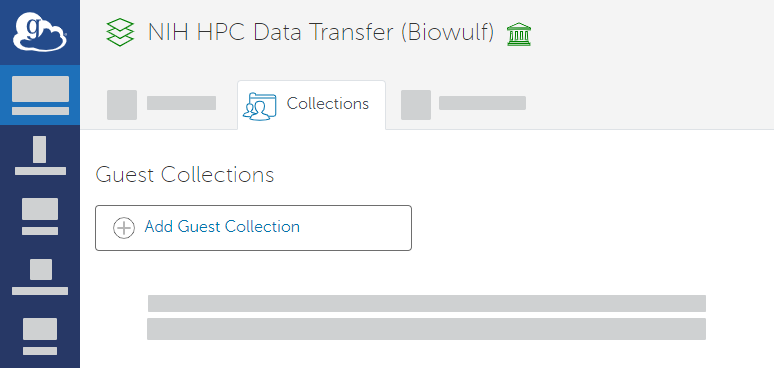
- In the search results, in the row for NIH HPC Data Transfer (Biowulf), click the collection details icon (). The details page appears. On the Collections tab, click Add Guest Collection.
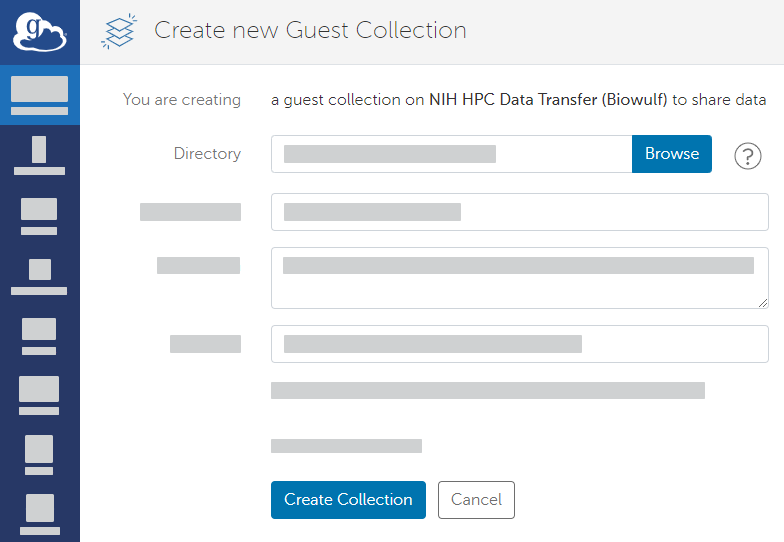
- On the Create new guest collection page, browse to or specify the path to the directory where you want to save your data. Click Create Guest Collection.
- In the Globus File Manager, search for NIH HPC Data Transfer (Biowulf). This is a public endpoint.
- Share your Globus endpoint:
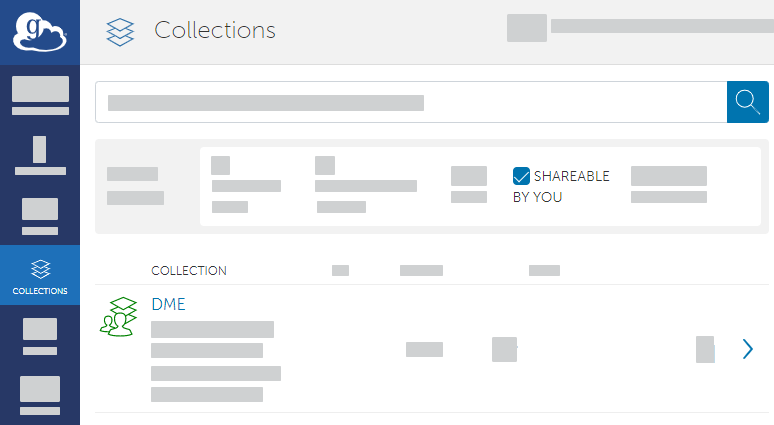
In the Globus File Manager, navigate to the list of collections by opening the side bar, clicking Collections, and then clicking Shareable by You.
Select your guest collection. Access the sharing configuration for that collection by clicking Permissions.
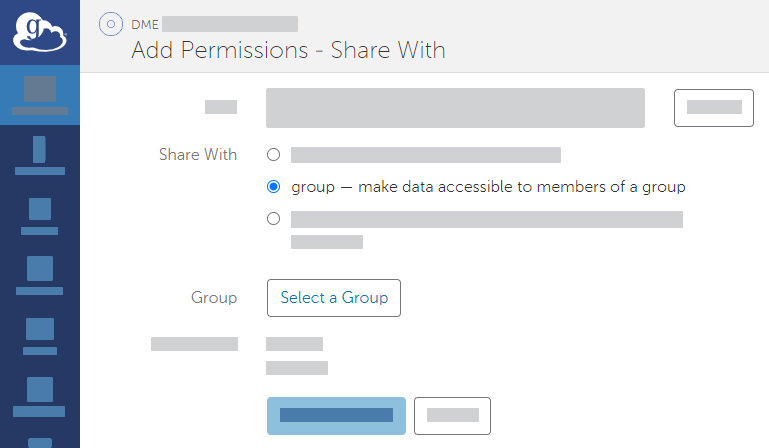
Add permissions by clicking Add Permissions - Share With. In the Share With field, select the option to share with a group. Click Select a Group.
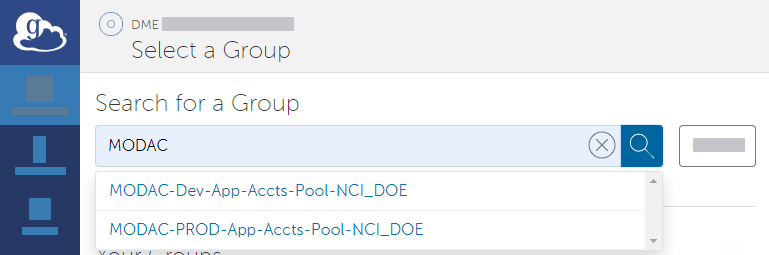
Search all groups for "HPCDME-" and select the HPCDME-PROD-App-Accts-Pool-FNLCR group.
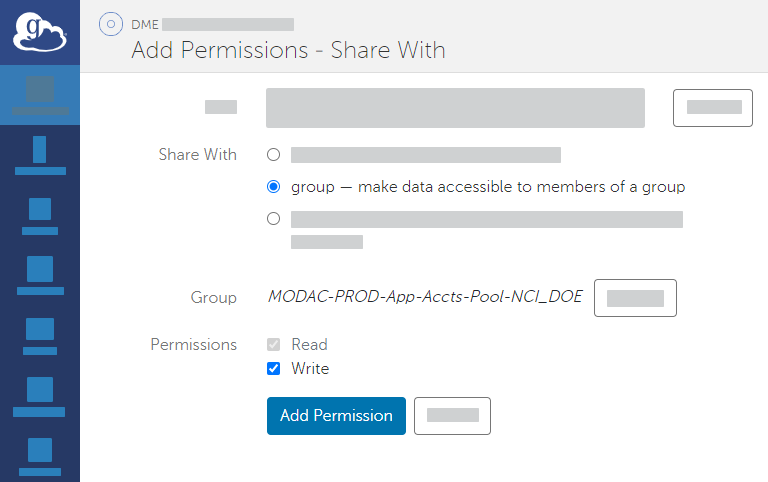
Panel borderColor black borderStyle solid Expand title Instructions to Use UAT If you intend to use the UAT tier of DME, select the HPCDME-DEV-App-Accts-Pool-FNLCR group. - Select the appropriate permission for the selected group: If you intend to download data from DME to the Globus endpoint, select the write permission.
- Click Add Permission. The system adds the HPCDME Globus group to the list for the current share.
Now you can register or download data in the GUI
The following activities in DME require a Globus endpoint:
- Registering multiple files, or files of more than 100 MB.
- Downloading a collection of data files.
- Downloading a single file asynchronously.
(TBD: I did these steps out of order, so I'm unable to see the process the way a new user would see it. Also, trying to find the EXE for the client update, so that I can right-click and run it with elevated permissions.)
To prepare an Globus endpoint:
- Send a request to NCI-Frederick Helpdesk for access to the nihfnlcr#gridftp1 Globus endpoint.
- When prompted to do so, complete the NCI at Frederick Access Request form.
- When prompted to do so, log in to Globus and install the Globus Connect Personal client, as described in Data transfer and sharing using Globus.
- Share the endpoint from your client to HPCDME, as described in Granting Write Access of Globus Endpoint to HPCDME. (Question: Section 9.9.1 of the user guide in GitHub has similar info. Should I draft a page in this wiki guide to replace this PDF?)
Now you can upload or download data as described in the following pages:
- Registering Data in Bulk from a Globus Endpoint via the Web GUI
- Downloading to a Single Data File Globus Endpoint via the Web GUIDownloading a Collection via the Web GUI
To register or download data via CLU, refer to the additional prerequisites described in Getting Started with DME CLU.