|
Page History
...
However, at this time, there is limited support for automatic evaluation of submitted results, results presentation, native support for medical images although many of these features are planned.
The HubZero (https://hubzero.org/) is an open source platform developed for scientific collaboration. It has been used heavily in a number of communities including nanoscience, earthquake engineering, molecular diagnostics and others. A version focused on cancer informatics can is hosted at nciphub.org. nciphub shares a lot of features with the Synapse platform. It allows user management and role-based access. Users can create groups that share common interest and collaborate within these groups. Files can be shared within projects. Other features include wiki, calendars, creating and sharing resources such as presentations, multimedia and even tools. Most common tools found on the various hubs are those based on simulations. Although nciphub has limited native support of medical imaging, libraries to handle medical images can be configured to work in the hub. Members of the Quantitative Imaging Network (QIN) are exploring the use of nciphub for challenges, especially for the communication and data sharing.
CodaLab (http://codalab.org/) is an open-source project that originated at Microsoft Research that was expressly created for hosting challenges and supporting reproducible research. The OuterCurve Foundation currently maintains it. Challenge organizers can easily set up challenges by creating a competition bundle that consists of data as well as evaluate tools. As part of the configuration files, the number of phases and duration (e.g. training, leaderboard, test) can be set up by the organizer. The evaluation program can be written in any language. Participants can upload results and get immediate feedback. The currently available version of CodaLab comes with scoring algorithms for image segmentation evaluation Organizers can extend the presentation of results to allow drilling down into the results with tables and charts. CodaLab currently uses the Azure platform although, in theory, it should be possible to deploy on other servers without a great deal of effort. CodaLab is also developing support for worksheets. These are resources to support reproducible research and for collaboration. Using these, researchers have compared a number of open source NLP tools on different public datasets http://research.microsoft.com/apps/video/default.aspx?id=225950 . As this technology continues to be developed, researchers will be able to quickly compare the performance of different algorithms on a range of datasets in the "cloud" by leveraging Azure technology.
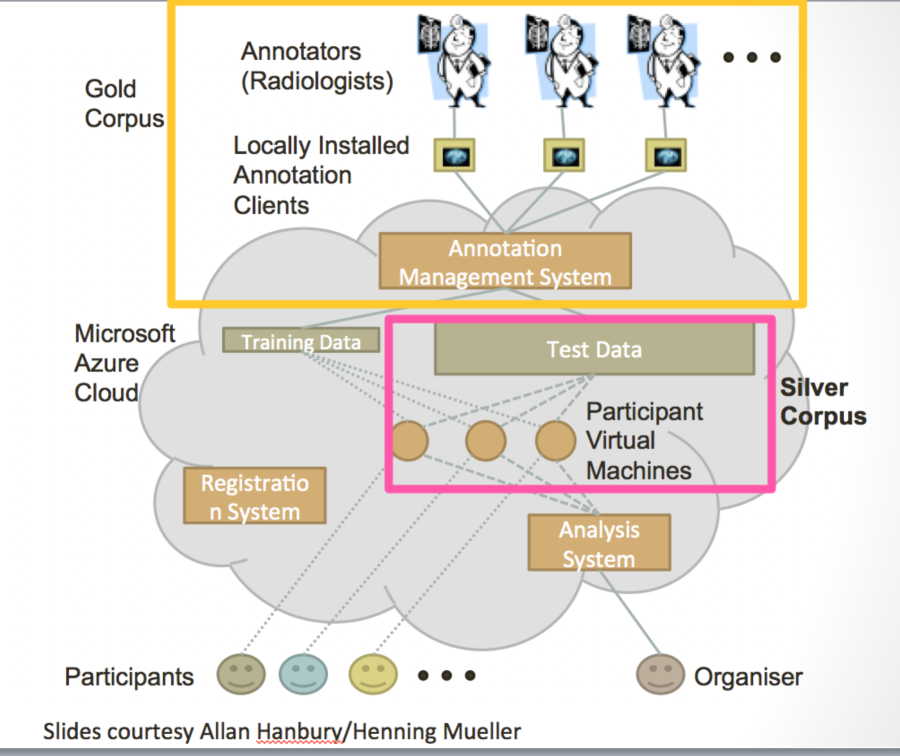
Visual Concept Extraction Challenge in Radiology (VISCERAL) is a large EU funded project to develop cloud-based challenge infrastructure. This open source platform, based on the Azure platform as described below, facilitates cloud-based challenges where the participants upload their algorithms rather than downloaded data and uploading algorithm output. This platform has been used for 4 medical imaging challenges at MICCAI and ISBI. Participants are provided virtual machines with access to the training data where they can deploy, configure and validate their algorithms. Once the training phase is completed, the virtual machines are then handed over to the organizers. The organizers can then run the algorithms on the test data. This feature, where the organizers and not the participants run the algorithms on the test data, is a unique to the VISCERAL system. This has a number of advantages in that the participants are never provided access to the test data, which reduces the risk of overfitting. Furthermore, it allows private data to be used for the testing phase and promotes unplanned dissemination of secure data. Finally, it supports the notion of reproducible research as the algorithms can always be rerun if the virtual machines are saved. Participants are allowed the share either source code or executables thus allowing both open and closed source algorithms to compete in the same venue.
Figure 8 Schematic diagrams of the VISCERAL system for cloud-based challenges
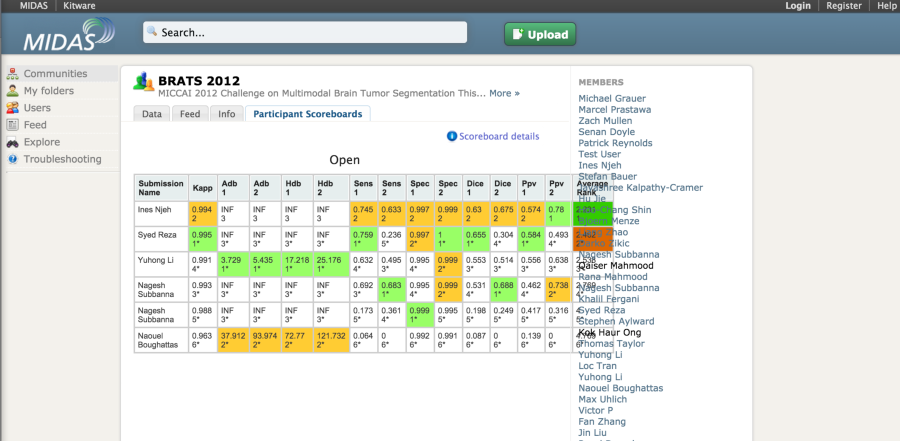
The MIDAS platform has been used to host a couple of imaging challenges. A special module is available to host challenges https://github.com/midasplatform/challenge . The developers of the platform also made available the COVALIC evaluation tool for segmentation challenges with the following metrics: Average distance of boundary surfaces, 95th percentile Hausdorff distance of boundary surfaces, Dice overlap, Cohen's kappa, Sensitivity, Specificity, Positive Predictive Value.
Once the participants have uploaded their submissions, the leaderboard updates the scores automatically.
A new version of the platform appears to be in development. http://challenge.kitware.com/#challenges/learn This system (COVALIC) is built on the TangeloHub platform--an open source data and analytics platform made up of three major components: Tangelo, Girder, and Romanesco.
Set this section apart from the review of the challenge management system.
Matrix of Features and Frameworks (1 -5*Some of the systems mentioned earlier are not listed in the matrix: Innocentive, TopCoder, ChallengePost, Midas/COVALIC)*
Below (in our opinion) is a table that rates the relative merits of the most relevant frameworks that we evaluated. The scale is 1-5 where 1 indicates excellent support for the feature while 5 indicates that that feature is not currently part of the system or there is limited support.
...