|
Page History
...
There were 3 sub-challenges within the radiology challenge. The primary goal of the radiology challenge was to perform segmentation from multimodal MRI of brain tumors. T1 (pre and post constrast), T2 and FLAIR MRI images were preprocessed (registered and resampled to 1mm isotropic) by the organizers and made available. Ground truth in the form of label maps (4 color –enhancing, necrosis, non-enhancing tumor and edema) were also provided for the training images in .mha format. Additional sub-tasks included longitudinal evaluation of the segmentations for patients who had imaging from multiple time points. Finally, the third subtask was to classify the tumors into one of the three classes (Low Grade II, Low Grade III, and High Grade IV glioblastoma multiforme (GBM)). However, sub-tasks 2 and 3 were primarily pushed out to future years.
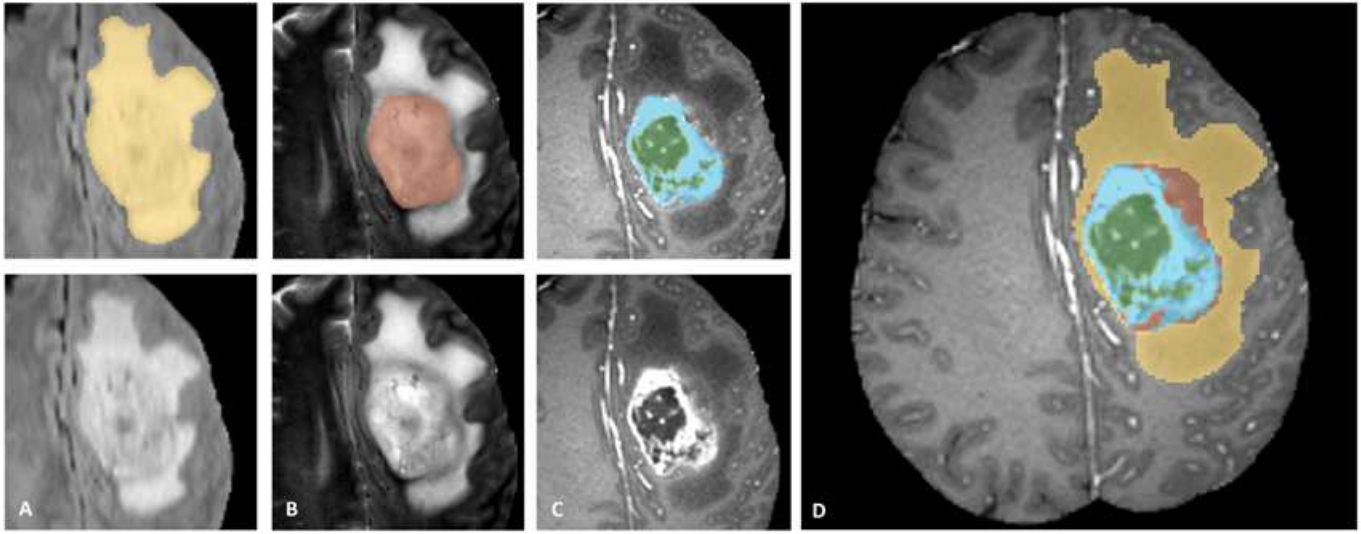
Figure 1. Label maps for the different sub-regions of the tumor used for the BraTS challenge. Manual annotation is performed by expert raters: the whole tumor visible in FLAIR (A), the tumor core visible in T2 (B), the enhancing active tumor visible in T1c (blue), surrounding the cystic/necrotic components of the core (green) (C). The segmentations are combined to generate the final labels (D): edema (yellow), non-enhancing solid core (red), active core (blue), non-solid core (green).
Pathology challenge also had classification and segmentation sub-tasks. The goal for the classification challenge was to classify the image into high grade and low-grade glioma while the goals for the segmentation challenge was to identify areas of necrosis.
...
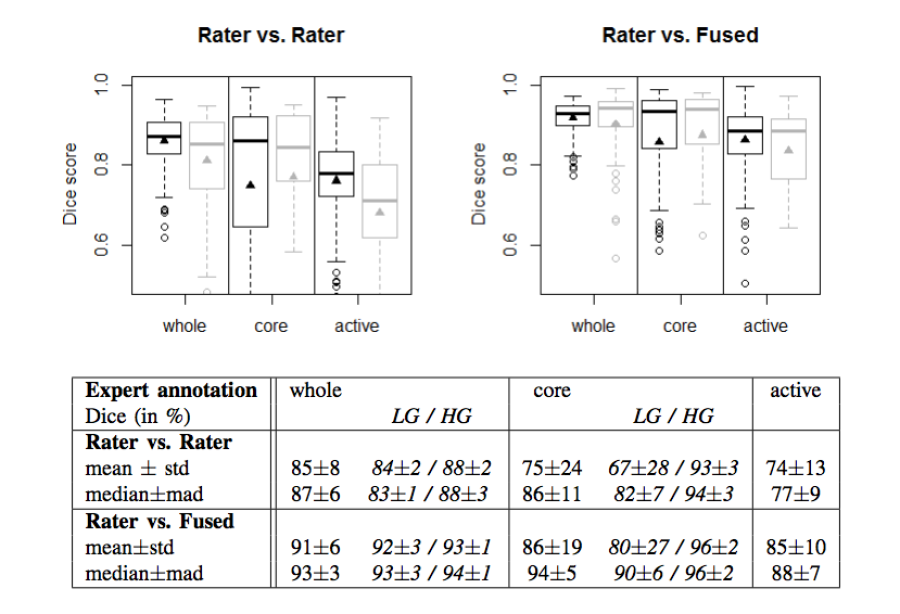
- The agreement between experts in not perfect (~0.8 Dice score)
- The agreement (between experts and between algorithms) is highest for the whole tumor and relatively poor for areas of necrosis and non-enhancing tumor
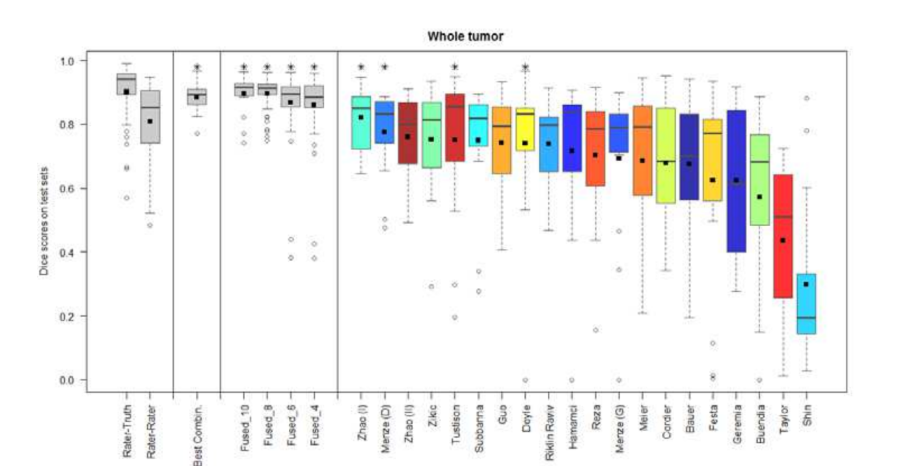
- Combining segmentations created by "best" algorithms created a segmentation that achieves overlap with consensus "expert" labels that approaches inter-rater overlap
- This approach can be used to automatically create large labeled datasets
- However, cases where this does not work and we still need to validate a subset of images with human experts
Figure 2. Dice coefficients of inter-rater agreement and of rater vs. fused label maps
Figure 3. Dice coefficients of individual algorithms and fused results indicating improvement with label fusion
More recently, the medical imaging community has begun organizing cloud-based challenges. The VISCERAL project (EU-funded effort) has organized a series of challenges at ISBI, MIICAI and other conferences where the participants in the challenge also share the algorithms and code, in addition to the results.
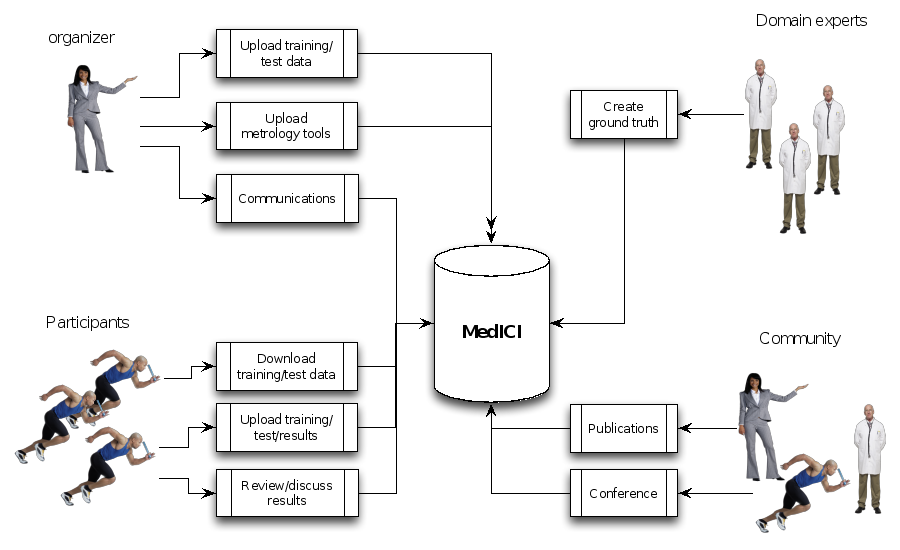
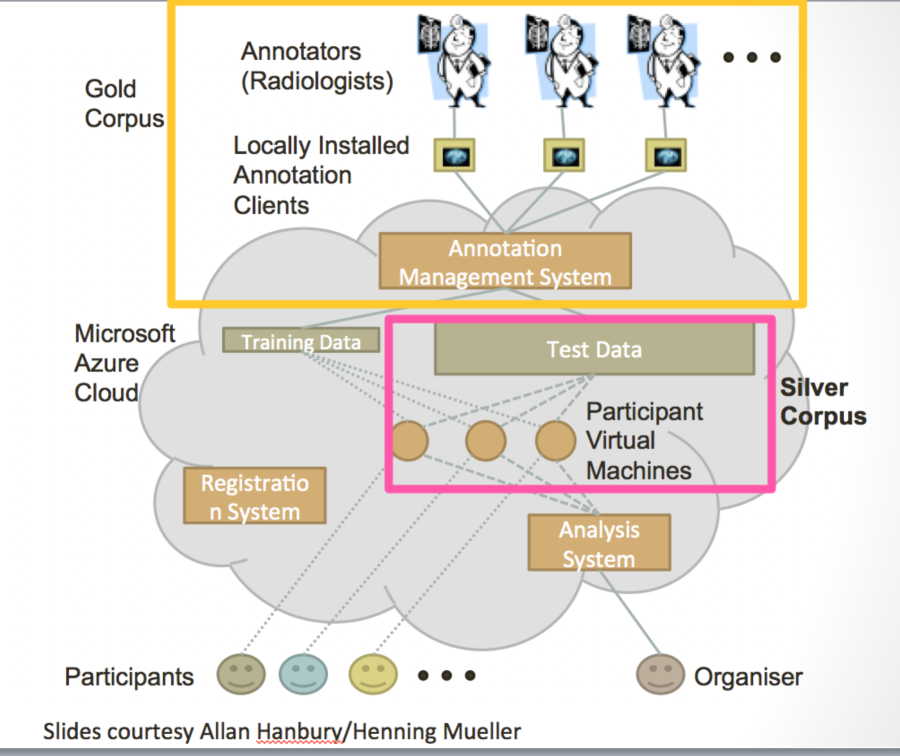
Below is a workflow diagram that describes the various stakeholders in the challenge and their tasks.
Figure 4. Challenge stakeholders and their tasks
Existing challenge infrastructure
...
- Average Precision
- Absolute Error
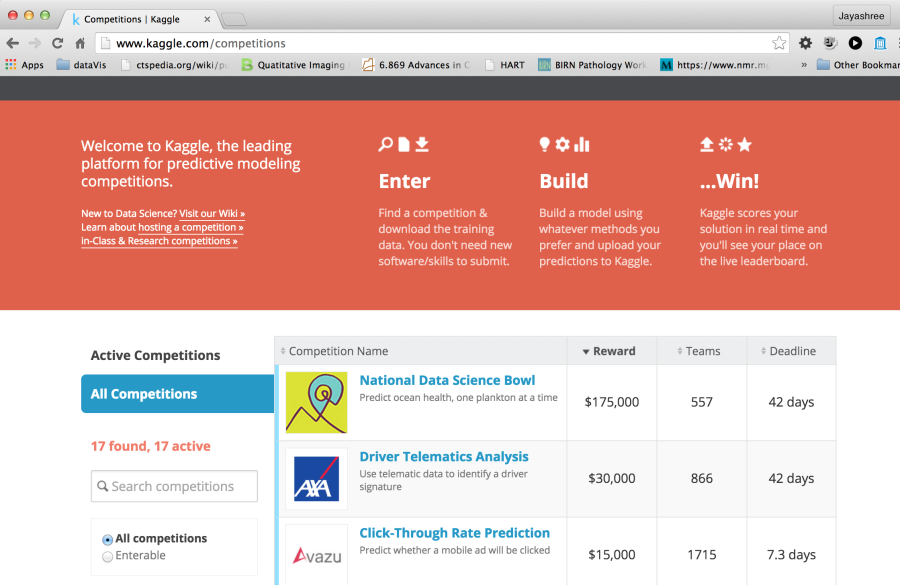
Figure 5. Portal for Kaggle, a leading website for challenges for data scientists
Topcoder is a similar popular website for software developers, graphic designers and data scientists. In this case, participants typically share their code or designs. They use the Appirio proprietary crowdsourcing development platform, built on Amazon Web Services, Cloud Foundry, Heroku, HTML5, Ruby and Java. A recent computational biology challenge run on Topcoder demonstrated that this crowdsourcing approach produced algorithmic solutions that greatly outperform commonly used algorithms such as BLAST for sequence annotation {Lakhani, 2013 #3789}. This competition was run with a $6000 prize and drew 733 participants (17% of whom submitted code) and the prize-winning algorithms were made available with an open source license.
...
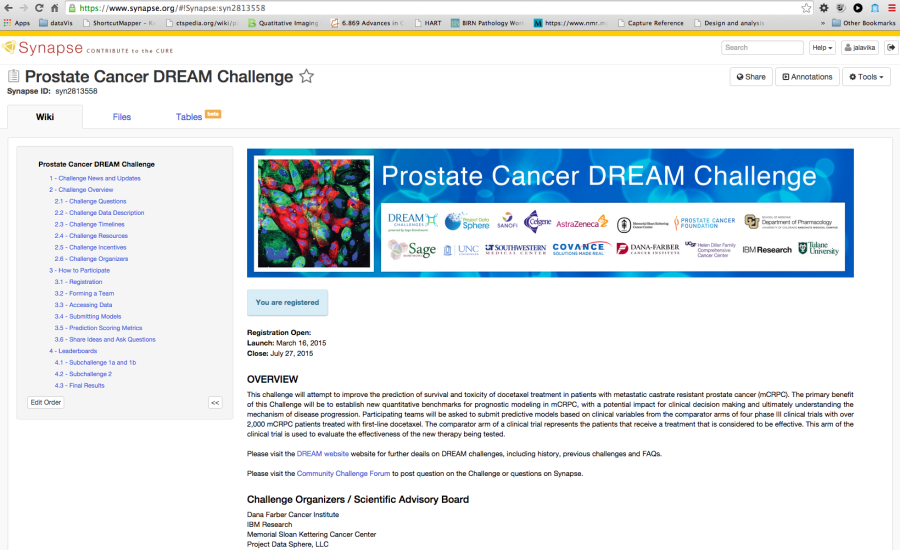
Synapse is both an open source platform and a hosted solution for challenges and collaborative activities created by Sage bionetworks. It has been used for a number of challenges including the DREAM challenge. Synapse allows the sharing of code as well as data. However, the code typically is in R, Python and similar languages. Synapse also has a nice programmatic interface and methods to upload/download data, submit results, create annotations and provenance through R, Python, command line and Java. These options can be configured for the different challenges. Content in Synapse is referenced by unique Synapse IDs. The three basic types of Synapse objects include projects, folders and files. These can be accessed through the web interface or through programmatic APIs. Experience and support for running image analysis code within Synapse is limited.
Figure 6. Portal for the Synapse platform
Figure 7. Example Challenge hosted in Synapse
COMIC framework is an open-source platform that facilitates the creation of challenges and has been used to host a number of medical imaging challenges. The Consortium for Open Medical Image Computing (COMIC) platform, built using Python/Django was created and is maintained by a consortium of five European medical image analysis groups including Radboud University, Erasmus, and UCL. They also offer a hosted site, with the hardware located at Fraunhofer MEVIS in Bremen, Germany. The current framework allows participants to create a website, add pages including wikis, create participant registrations, methods for organizers to upload data and participants to download data (for instance through Dropbox). However, the platform including ways to visualize medical data and results is still under development as are options to share algorithms and perform challenges in the cloud.
...
Visual Concept Extraction Challenge in Radiology (VISCERAL) is a large EU funded project to develop cloud-based challenge infrastructure. This open source platform, based on the Azure platform as described below, facilitates cloud-based challenges where the participants upload their algorithms rather than downloaded data and uploading algorithm output. This platform has been used for 4 medical imaging challenges at MICCAI and ISBI. Participants are provided virtual machines with access to the training data where they can deploy, configure and validate their algorithms. Once the training phase is completed, the virtual machines are then handed over to the organizers. The organizers can then run the algorithms on the test data. This feature, where the organizers and not the participants run the algorithms on the test data, is a unique to the VISCERAL system. This has a number of advantages in that the participants are never provided access to the test data, which reduces the risk of overfitting. Furthermore, it allows private data to be used for the testing phase and promotes unplanned dissemination of secure data. Finally, it supports the notion of reproducible research as the algorithms can always be rerun if the virtual machines are saved. Participants are allowed the share either source code or executables thus allowing both open and closed source algorithms to compete in the same venue.
Figure 8. Schematic diagrams of the VISCERAL system for cloud-based challenges
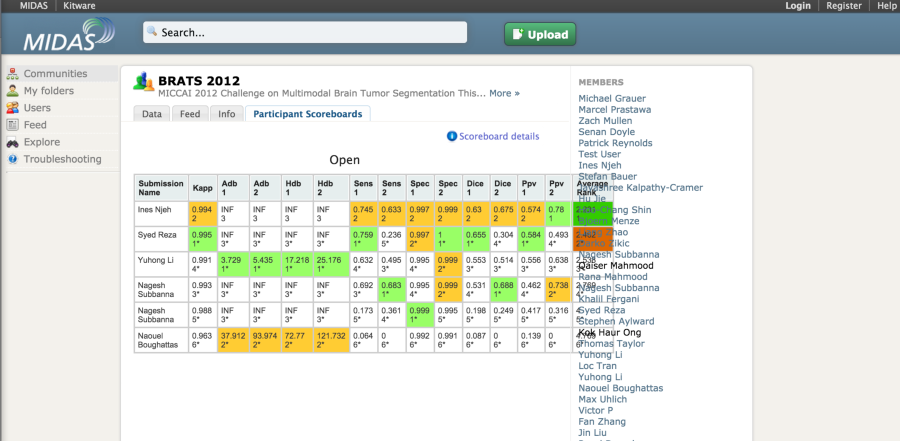
The MIDAS platform has been used to host a couple of imaging challenges. A special module is available to host challenges. The developers of the platform also made available the COVALIC evaluation tool for segmentation challenges with the following metrics: Average distance of boundary surfaces, 95th percentile Hausdorff distance of boundary surfaces, Dice overlap, Cohen's kappa, Sensitivity, Specificity, Positive Predictive Value.
Once the participants have uploaded their submissions, the leaderboard updates the scores automatically.
A new version of the platform appears to be in development. This system (COVALIC) is built on the TangeloHub platform--an open source data and analytics platform made up of three major components: Tangelo, Girder, and Romanesco.
Matrix of Features and Frameworks (1 -5)
...
- Shi S, Pei J, Sadreyev RI, Kinch LN, Majumdar I, Tong J, Cheng H, Kim BH, Grishin NV. Analysis of CASP8 targets, predictions and assessment methods. Database : the journal of biological databases and curation. 2009;2009:bap003. doi: 10.1093/database/bap003. PubMed PMID: 20157476; PubMed Central PMCID: PMC2794793.
- Brownstein CA, Beggs AH, Homer N, Merriman B, Yu TW, Flannery KC, DeChene ET, Towne MC, Savage SK, Price EN, Holm IA, Luquette LJ, Lyon E, Majzoub J, Neupert P, McCallie D, Jr., Szolovits P, Willard HF, Mendelsohn NJ, Temme R, Finkel RS, Yum SW, Medne L, Sunyaev SR, Adzhubey I, Cassa CA, de Bakker PI, Duzkale H, Dworzynski P, Fairbrother W, Francioli L, Funke BH, Giovanni MA, Handsaker RE, Lage K, Lebo MS, Lek M, Leshchiner I, MacArthur DG, McLaughlin HM, Murray MF, Pers TH, Polak PP, Raychaudhuri S, Rehm HL, Soemedi R, Stitziel NO, Vestecka S, Supper J, Gugenmus C, Klocke B, Hahn A, Schubach M, Menzel M, Biskup S, Freisinger P, Deng M, Braun M, Perner S, Smith RJ, Andorf JL, Huang J, Ryckman K, Sheffield VC, Stone EM, Bair T, Black-Ziegelbein EA, Braun TA, Darbro B, DeLuca AP, Kolbe DL, Scheetz TE, Shearer AE, Sompallae R, Wang K, Bassuk AG, Edens E, Mathews K, Moore SA, Shchelochkov OA, Trapane P, Bossler A, Campbell CA, Heusel JW, Kwitek A, Maga T, Panzer K, Wassink T, Van Daele D, Azaiez H, Booth K, Meyer N, Segal MM, Williams MS, Tromp G, White P, Corsmeier D, Fitzgerald-Butt S, Herman G, Lamb-Thrush D, McBride KL, Newsom D, Pierson CR, Rakowsky AT, Maver A, Lovrecic L, Palandacic A, Peterlin B, Torkamani A, Wedell A, Huss M, Alexeyenko A, Lindvall JM, Magnusson M, Nilsson D, Stranneheim H, Taylan F, Gilissen C, Hoischen A, van Bon B, Yntema H, Nelen M, Zhang W, Sager J, Zhang L, Blair K, Kural D, Cariaso M, Lennon GG, Javed A, Agrawal S, Ng PC, Sandhu KS, Krishna S, Veeramachaneni V, Isakov O, Halperin E, Friedman E, Shomron N, Glusman G, Roach JC, Caballero J, Cox HC, Mauldin D, Ament SA, Rowen L, Richards DR, San Lucas FA, Gonzalez-Garay ML, Caskey CT, Bai Y, Huang Y, Fang F, Zhang Y, Wang Z, Barrera J, Garcia-Lobo JM, Gonzalez-Lamuno D, Llorca J, Rodriguez MC, Varela I, Reese MG, De La Vega FM, Kiruluta E, Cargill M, Hart RK, Sorenson JM, Lyon GJ, Stevenson DA, Bray BE, Moore BM, Eilbeck K, Yandell M, Zhao H, Hou L, Chen X, Yan X, Chen M, Li C, Yang C, Gunel M, Li P, Kong Y, Alexander AC, Albertyn ZI, Boycott KM, Bulman DE, Gordon PM, Innes AM, Knoppers BM, Majewski J, Marshall CR, Parboosingh JS, Sawyer SL, Samuels ME, Schwartzentruber J, Kohane IS, Margulies DM. An international effort towards developing standards for best practices in analysis, interpretation and reporting of clinical genome sequencing results in the CLARITY Challenge. Genome biology. 2014;15(3):R53. doi: 10.1186/gb-2014-15-3-r53. PubMed PMID: 24667040; PubMed Central PMCID: PMC4073084.
- Omberg L, Ellrott K, Yuan Y, Kandoth C, Wong C, Kellen MR, Friend SH, Stuart J, Liang H, Margolin AA. Enabling transparent and collaborative computational analysis of 12 tumor types within The Cancer Genome Atlas. Nature genetics. 2013;45(10):1121-6. doi: 10.1038/ng.2761. PubMed PMID: 24071850; PubMed Central PMCID: PMC3950337.
- Abdallah K, Hugh-Jones C, Norman T, Friend S, Stolovitzky G. The Prostate Cancer DREAM Challenge: A Community-Wide Effort to Use Open Clinical Trial Data for the Quantitative Prediction of Outcomes in Metastatic Prostate Cancer. The oncologist. 2015. doi: 10.1634/theoncologist.2015-0054. PubMed PMID: 25777346.
- Jarchum I, Jones S. DREAMing of benchmarks. Nat Biotechnol. 2015;33(1):49-50. doi: 10.1038/nbt.3115. PubMed PMID: 25574639.