|
Page History
...
| MICCAI 2015 Challenges | Description | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Combined Radiology and Pathology Classification | The datasets for this challenge are Radiology and Pathology images obtained from the same patients. Each case corresponds to a single patient. There is one Radiology image and one whole slide tissue image for each case. The training set contains a total of 32 cases: 16 cases that are classified by pathologists as Oligodendroglioma and 16 cases classified as Astrocytoma. The test set will have 20 cases. Please note that the number of cases in the test set may not be equally partitioned between the two sub-types. The whole slide tissue images are stored in Aperio SVS format. There are open source tools and libraries that can read these images: OpenSlide
| |||||||||||||||||||
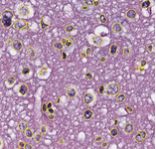
Segmentation of Nuclei in Pathology Images | The goal of this challenge is to evaluate the performance of algorithms for detection and segmentation of nuclear material in a tissue image. Participants are asked to detect and segment all the nuclei in a given set of image tiles extracted from whole slide tissue images. The algorithm results will be compared with consensus pathologist-segmented sub-regions. Winners will be ranked based on their nuclei segmentation best matching the reference standards. The reference standard for the challenge will be pathologist-generated nuclear segmentation on select regions of TCGA Glioma whole slide images. |
...