|
Page History
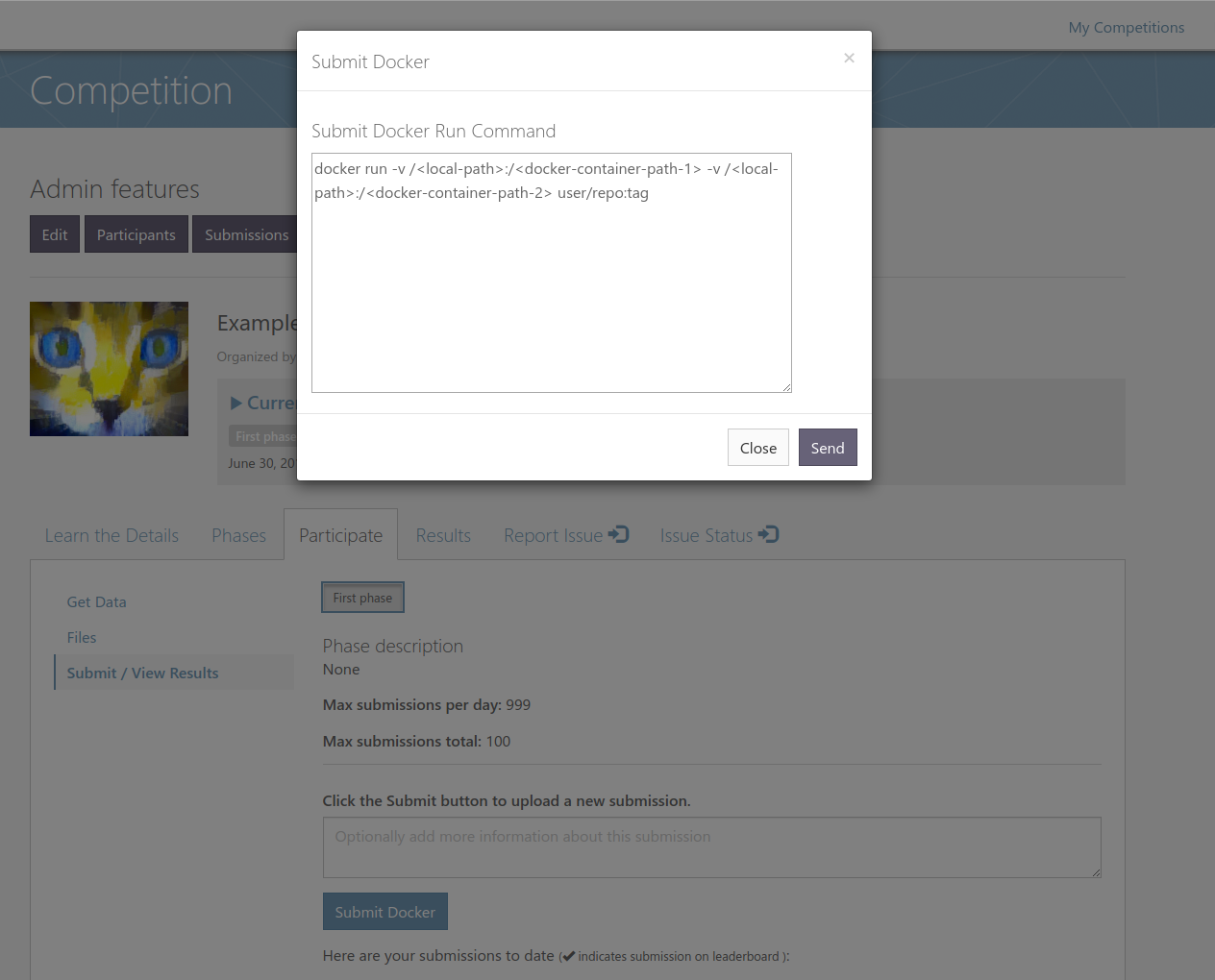
1) Talked with NCI, Spyridon and Tahsin and discussed hardware requirements and things to keep in mind for future challengesRe-implemented docker submissions for MedICI site (should work for upcoming challenges).
2) Got a tutorial for ePAD and learned how to make annotations and save them to the DB.
3) Also learned to download TCIA images and add them to ePAD.
| Planned for next month |
|---|
1) |
Fine tune code changes. 2) |
QA implementation by re-creating MICCAI 2019 challenge and submitting dockers to test implementation. |
| Comments |
|---|
Things that went well: |
We ran the dockers successfully.
Do we want to continue using docker containers? Yes
Participants were able to submit their resultant *.csv files relatively easily.
Things that could be improved: |
The participant dockers seemed to be able to delete a couple of images. I think participants shouldn’t be able to adjust files their containers are mounted to. This might be as simple as read-only permissions. Cuong says we can mount files to dockers with read-only permissions.
We should finalize metrics for the evaluate.py script so that the final leaderboard columns are as we expected.
We should know if we will need to use shm (shared memory size) before hand:
$ sudo docker run --runtime=nvidia --shm-size 50G -v /home/bbearce/input_images:/data/images -v /home/bbearce/yunzhe_docker_run/output_result:/data/output xyzacademic/njit_datasci:cpmv
Remember to delete the index.html files that come with a curl or wget command. This caused some errors and confusion for participants.
Docker worker container needs updated python for certain scikit-learn methods.
Is there any way to check if a team has multiple screen names or accounts ahead of time?
FTP:
Server on same VM as CodaLab.
Have data available all year so participants have more time.
|
| Meeting | Date |
|---|---|
| Bi-weekly Meeting #1 | 14 Nov |
| Bi-weekly Meeting #2 | 28 Nov |
Milestones
| Description | Date |
|---|
| Successfully got a docker run command to a worker VM and executed it. | 01/06 |
| /2019 |
| Learned enough about ePAD to setup ground truth annotation sets for professional annotators. |
Task
| Description | Resolution | Status | Creation Date | Close Date |
|---|---|---|---|---|
| Figure out how to run a docker through CodaLab |
| Complete |
| Done | 10/09/2019 |
| 01/06/2019 | |||
| Finish ePAD tutorials (they are online)..there is always more to learn | In Progress | In progress | 10/09/2019 |
| -- | ||||
| Contact Ryan Birmingham for caMicroscope tutorial | In Progress | Coordinating with Ryan | 10/09/2019 | -- |
Risks
| Description | Mitigation | Rank | Status | Creation Date | Realization | Close Date |
|---|
| The worker queue system in theory can handle multiple submissions by participants. Should that assumption not hold we could be in for a nasty surprise that this doesn't work as expected. | Test real AI containers that can take up to half a day to run on the system and submit multiple containers. | 1 | In Progress | 1/13/2020 | 1/13/2020 |
| -- |