|
Page History
<?xml version="1.0" encoding="utf-8"?>
<html>
<span style="font-size:medium;">
<strong>Applicable Applicable Releases:</strong> caArray 2.X
<strong>Date entered:</strong> June 30, 2010
</span>
caArray Usage
{{question}}Question: What is the difference between the annotation options that are available when importing data?
...
We were manually uploading CHP and CEL files (as well as supplemental files) for 72 patients into our instance of caArray. When we were importing, caArray offered 4 annotation options. What is the difference between these options?
<noinclude>
\ width="100%" style="border: solid 1px #A3B1BF; background: #F5FAFF" |
...
| Scrollbar | ||
|---|---|---|
|
| Page info | ||||
|---|---|---|---|---|
|
| Composition Setup |
|---|
cloak.toggle.type = text
cloak.toggle.open=[show]
cloak.toggle.close=[hide]
|
| Panel | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
|
Topic:
Release:
Date entered:
Question
Answer
Have a comment?
Please leave your comment in the caArray End User Forum.
| Scrollbar | ||
|---|---|---|
|
answer}}Answer:{{answerEnd
The import process is the last step of data uploading in caArray, which allows the array data to be stored in the database. During the importing process, caArray associates the data with the appropriate biomaterial and hybridization annotation by creating an annotation chain of source > sample > extract > labeled extract > hybridization. The array data can be downloaded only if the data were associated properly.
Import MAGE-TAB set
If a MAGE-TAB set (IDF and SDRF) is imported along with the data files, where the SDRF file refers to each of the data files, caArray will use the information provided in SDRF to determine how to create sources, samples, extracts, labeled extracts and hybridizations.
Import Only the datafile
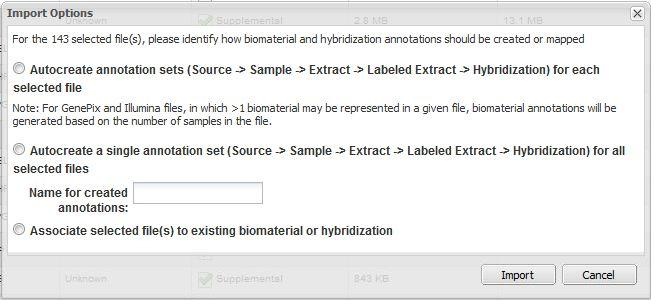
If only data files (for example, .cel, .chp, etc.) are imported, caArray offers three options to associate the data and annotation (Figure 1):
Option 1. Autocreate annotation sets ... for each selected file
For every unique file name to be imported, caArray automatically creates a Source - Sample - Extract - Labeled Extract - Hybridization chain corresponding to each data file imported.
Option 2. Autocreate a single annotation set ... for all selected files
caArray creates a single Source - Sample - Extract - Labeled Extract - Hybridization chain, and associates all selected data files with this single chain.
Option 3. Associate selected file(s) to existing biomaterial or hybridization
caArray displays all available sources, samples, extracts, labeled extracts and hybridizations. User select one of these, caArray associated the selected files with that biomaterial or hybridization. Note that additional items in the chain (to the right of the selected biomaterial) may need to be generated by the System.
Trouble Shooting: Choose the right option
The proper association between the array data and annotation data is critical to a successful array data import.
...