|
Page History
...
This chapter describes how to use caIntegrator tools to analyze data in subject annotation or genomic studies that have been deployed in caIntegrator.
Topics in this chaptersection chapter include the following:
| Table of Contents | ||
|---|---|---|
|
...
- Select the study whose data you want to analyze in the upper right portion of the caIntegrator page. You must select a study saved as a subject annotation study, but which has genomic data.
- Click GenePattern Analysis in the left sidebar of caIntegrator. This opens the GenePattern Analysis Status page.
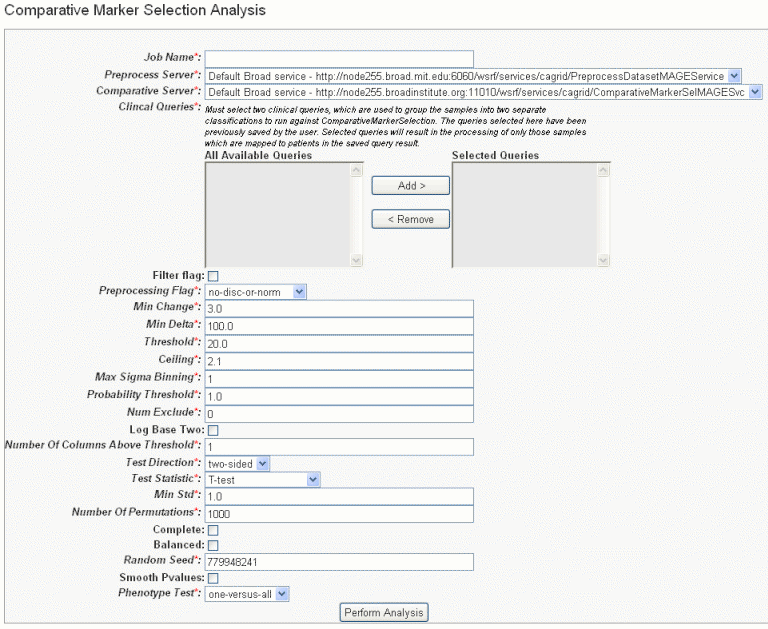
- In the GenePattern Analysis Status page, select Comparative Marker Selection (Grid Service) from the drop down list and click New Analysis Job. This opens the Comparative Marker Selection Analysis page ().
- Select or define CMS analysis parameters, described in the following table. An asterisk indicates required fields. The default settings are valid; they should provide valid results.
CMS Parameter
Description
Job Name*
Assign a unique name to the analysis you are configuring.
Preprocess Server*
A server which hosts the grid-enabled data GenePattern PreProcess Dataset module. Select one from the list and caIntegrator will use the selected server for this portion of the processing.
Comparative Server*
A server which hosts the grid-enabled data GenePattern Comparative Marker Selection module. Select one from the list and caIntegrator will use the selected server for this portion of the processing.
Annotation Queries and Lists*
All subject annotation queries and gene lists with appropriate data for the analysis are listed. Select and move two or more queries from the All Available Queries panel to the Selected Queries panel using the Add > and Remove < buttons.
<ac:structured-macro ac:name="unmigrated-wiki-markup" ac:schema-version="1" ac:macro-id="674c2963b4890f60-480b79b6-47dd4a6c-a3658811-c90711c5094515f1ce3b92ee"><ac:plain-text-body><![CDATA[Note: The [SL] and [Q] prefixes to list names indicate "Subject Lists" or "Saved Queries". A "G" in the prefix indicates the list is Global. For more information, see on page 69.]]></ac:plain-text-body></ac:structured-macro>
Filter Flag
Variation filter and thresholding flag
Preprocessing Flag*
Discretization and normalization flag
Min Change*
Minimum fold change for filter
Min Delta*
Minimum delta for filter
Threshold*
Value for threshold
Ceiling*
Value for ceiling
Max Sigma Binning*
Maximum sigma for binning
Probability Threshold*
Value for uniform probability threshold filter
Num Exclude*
Number of experiments to exclude (max & min) before applying variation filter
Log Base Two
Whether to take the log base two after thresholding; default setting is "Yes".
Number of Columns Above Threshold*
Remove row if n columns are not >= than the given threshold
In other words, the module can remove rows in which the given number of columns does not contain a value greater or equal to a user defined threshold.Test Direction*
The test to perform (up-regulated for class0; up-regulated for class1, two sided). By default, Comparative Marker Selection performs the two-sided test.
Test Statistic*
Select the statistic to use.
Min Std*
The minimum standard deviation if test statistic includes the min std option. Used only if test statistic includes the min std option.
Number of Permutations*
The number of permutations to perform. (Use 0 to calculate asymptotic P-values.) The number of permutations you specify depends on the number of hypotheses being tested and the significance level that you want to achieve (3). The greater the number of permutations, the more accurate the P-value.
Complete – Perform all possible permutations. By default, complete is set to No and Number of Permutations determines the number of permutations performed. If you have a small number of samples, you might want to perform all possible permutations.
Balanced – Perform balanced permutationsRandom Seed*
The seed for the random number generator.
Smooth P-values
Whether to smooth P-values by using the Laplace's Rule of Succession. By default, Smooth P-values is set to Yes, which means P-values are always less than 1.0 and greater than 0.0.
Phenotype Test*
Tests to perform when class membership has more than 2 classes: one versus-all, all pairs.
Note: The P-values obtained from the one-versus-all comparison are not fully corrected for multiple hypothesis testing.- Comparative Marker Selection analysis options
Anchor RTF31353237303a205461626c65 RTF31353237303a205461626c65
- Comparative Marker Selection analysis options
- When you have completed the form, click Perform Analysis.
caIntegrator takes you to the JobStatus/Launch page where you will see the job and its status in the Status column of the list ().
- When the job is complete, the system displays a completion date on the GenePattern Analysis status page. Click the Download link. This downloads zipped result files to your local work station. The number of files and their file type will vary according to the processing. The results format is compatible with GenePattern visualizers and can be uploaded within GenePattern.
...
- Select the study whose data you want to analyze in the upper right portion of the caIntegrator page. You must select a study with gene expression data.
- Click GenePattern Analysis in the left sidebar of caIntegrator. This opens the GenePattern Analysis Status page.
- In the GenePattern Analysis Status page, select Principal Component Analysis (Grid Service) from the drop down list and click New Analysis Job. This opens the Principal Component Analysis page ().
- Select or define PCA analysis parameters, described in the following table. You must enter a job name and select an annotation query, but you can accept the other default settings..
PCA Parameters
Description
Job Name*
Assign a unique name to the analysis you are configuring.
Principal Component Analysis Server*
A server which hosts the grid-enabled data GenePattern Principal Component Analysis module. Select one from the list and caIntegrator will use the selected server for this portion of the processing.
Annotation Queries*
All annotation queries display in this list. Select one or more of these queries to define which samples are analyzed using PCA. If you select more than one query, then the union of the samples returned by the multiple queries is analyzed.
Cluster By*
Selecting rows looks for principal components across all expression values, and selecting columns looks for principal components across all samples.
- PCA analysis options
Anchor RTF39353234343a205461626c65 RTF39353234343a205461626c65
- PCA analysis options
- If you want to preprocess the data set, click Enable the Preprocess Dataset. This opens an additional set of parameters (), discussed in the following table . The preprocessing is executed prior to running the PCA.
PCA Preprocessing Parameters
Description
Preprocess Server*
A server which hosts the grid-enabled data GenePattern PreProcess Dataset module. Select one from the list and caIntegrator will use the selected server for this portion of the processing.
Filter Flag
Variation filter and thresholding flag
Preprocessing Flag
Discretization and normalization flag
Min Change
Minimum fold change for filter
Min Delta
Minimum delta for filter
Threshold
Value for threshold
Ceiling
Value for ceiling
Max Sigma Binning
Maximum sigma for binning
Probability Threshold
Value for uniform probability threshold filter
Num Exclude
Number of experiments to exclude (max & min) before applying variation filter
Log Base Two
Whether to take the log base two after thresholding
Number of Columns Above Threshold
Remove row if n columns no >= than the given threshold
Parameters for preprocessing data sets for PCAAnchor RTF36393835383a205461626c65 RTF36393835383a205461626c65
- When you have completed the form, click Perform Analysis.
- When the job is complete, the system displays a completion date on the GenePattern Analysis status page. Click the Download link. This downloads zipped result files to your local work station. The number of files and their file type will vary according to the processing. The results format is compatible with GenePattern visualizers and can be uploaded within GenePattern.
...
- Select the study whose data you want to analyze in the upper right portion of the caIntegrator page. You must select a study with copy number (either Affymetrix SNP or Agilent Copy Number) data.
- Click GenePattern Analysis in the left sidebar of caIntegrator. This opens the GenePattern Analysis Status page.
- In the GenePattern Analysis Status page, select GISTIC (Grid Service) from the drop down list and click New Analysis Job. This opens the GISTIC Analysis page ().
- Select or define GISTIC analysis parameters, as described in the following table. You must indicate a Job Name, but you can accept the other defaults settings, which are valid and should produce valid results.
GISTIC Parameters
Description
Job Name*
Assign a unique name to the analysis you are configuring.
GISTIC Service Type*
Select whether to use the GISTIC web service or grid service and provide or select the service address. If the web service is selected, authentication information is also required
GenePattern User Name/Password
Include these to log into GenePattern for the analysis.
Annotation Queries and Lists
All annotation queries display in this list as well as an option to select all non-control samples. Select an annotation query if you wish to run GISTIC on a subset of the data and select all non-control samples if wish to include all samples.
Select Platform
This option appears only if more than one copy number platform exists in the study. Select the appropriate platform from the drop-down list ().
Exclude Sample Control Set
From the drop-down list, select the name of the control set you want to exclude from the analysis. Click None if that is applicable.
Amplifications Threshold*
Threshold for copy number amplifications. Regions with a log2 ratio above this value are considered amplified. Default = 0.1.
Deletions Threshold*
Threshold for copy number deletions. Regions with a log2 ratio below the negative of this value are considered deletions. Default = 0.1.
Join Segment Size*
Smallest number of markers to allow in segments from the segmented data. Segments that contain fewer than this number of markers are joined to the neighboring segment that is closest in copy number. Default = 4.
<ac:structured-macro ac:name="unmigrated-wiki-markup" ac:schema-version="1" ac:macro-id="20eedb88986d3945-028c8e32-4f474352-85cca4f0-acde89800359ba309a589bec"><ac:plain-text-body><![CDATA[
QV Thresh[hold]*
Threshold for q-values. Regions with q-values below this number are considered significant. Default = 0.25.
]]></ac:plain-text-body></ac:structured-macro>
Remove X*
Flag indicating whether to remove data from the X-chromosome before analysis. Allowed values = {1,0}. Default = 1(yes).
cnv File
This selection is optional.
Browse for the file. There are two options for the CNV file.
Option #1 enables you to identify CNVs by marker name. Permissible file format is described as follows:
A two column, tab-delimited file with an optional header row. The marker names given in this file must match the marker names given in the markers_file. The CNV identifiers are for user use and can be arbitrary. The column headers are: - Marker Name
- CNV Identifier
Option #2 enables you to identify CNVs by genomic location. Permissible file format is described as follows:
A 6 column, tab-delimited file with an optional header row. The 'CNV Identifier', 'Narrow Region Start' and 'Narrow Region End' are for user use and can be arbitrary. The column headers are: - CNV Identifier
- Chromosome
- Narrow Region Start
- Narrow Region End
- Wide Region Start
- Wide Region End|
- GISTIC analysis parameters
- When you have completed the form, click Perform Analysis.
- When the job is complete, the system displays a completion date on the GenePattern Analysis status page. Click the Download link. This downloads zipped result files to your local work station. The number of files and their file type will vary according to the processing. The results format is compatible with GenePattern visualizers and can be uploaded within GenePattern.
- Additionally, upon completion of a successful GISTIC anaylsis, caIntegrator automatically displays the two gene lists that it generates in the Gene List Picker so that you can use them in a caIntegrator query or plot calculation. The lists are visible only to your userID. For more information, see . The genes will also display in Saved Copy Number Analyses in the left sidebar. See on page 74.
- If samples from a copy number source are deleted, the GISTIC job in which they are appear is also deleted.
...
- With the appropriate study open, at the bottom of the Query Results page, click the View in Integrative Genomics Viewer button.
- If you click the button at the bottom of the page with any of the query results line items selected, caIntegrator creates IGV files, with a monitor informing you of this. After the files are created, click the Launch Integrative Viewer hypertext link.
Follow the instructions through the intermediate dialog boxes. After clicking Open with the Java program listed, the opens, displaying the dataset in the computer screen shown as follows ().Anchor RTF31393435313a204e756d6265 RTF31393435313a204e756d6265
- Move your mouse to hover over the genes graphic at the bottom of the page, indicated in the figure .
- Click the mouse when you've identified a gene of interest.
This opens the genome site at UCSC , where you can learn more about the gene ().
Go to the following website for a user guide for IGV: Method 2
- With the appropriate study open, click Integrative Genomics Viewer on the left sidebar.
- This opens the View IGV Selector page ().
- In the drop-down list, select the Gene Expression Platform for the data you want to view.
- Select the Copy Number Platform ID.
- The Annotations - Default panel displays existing annotation fields for the gene expression data in the open study. Select those fields you want to view when you open the IGV. Use the buttons for convenience if you want to Select All or Unselect All, when all are checked.
- Click View to see the data in the Integrative Genomic Viewer. caIntegrator creates IGV files of the data.
- After the files are created, click the Launch Integrative Viewer hypertext link that appears.
- Continue with .
...
- With the appropriate study open, at the bottom of the Query Results page, click the View in Heat Map Viewer button.
- If you click the button at the bottom of the page with any of the query results line items selected, caIntegrator creates HMV files, with a monitor informing you of this. After the files are created, click the Launch Heat Map Viewer hypertext link.
Follow the instructions through the intermediate dialog boxes. After clicking Open with the Java program listed, the runs, displaying the dataset in the computer screen, shown as follows ().Anchor RTF39333030383a204e756d6265 RTF39333030383a204e756d6265
Go to the following website for Heat Map Viewer documentation: Method 2
- With the appropriate study open, click Heat Map Viewer on the left sidebar.
- This opens the View Heat Map Viewer Selector page ().
- Select the appropriate Copy Number Platform in the drop down list.
- The Annotations - Default panel displays existing annotation fields for the gene expression data in the open study. Select one or more annotations in the annotation list. For convenience, you can use the Select All or Unselect All buttons.
- Click View to view the data you select in Heat Map Viewer. caIntegrator creates Heat Map Viewer files of the data.
- After the files are created, click the Launch Heat Map Viewer hypertext link that appears.
- Continue with .
- For interpretation of the results and using HMV features, see the help files opened from HMV.
...