|
Page History
| Scrollbar | ||
|---|---|---|
|
...
| Page info | ||||
|---|---|---|---|---|
|
Version 1.0.2
...
This installation guide outlines the supported configurations and technical installation instructions for LexGrid Vocabulary Services for caBIG TMcaBIG©, referred to as LexBIG for the remainder of the guide. Directions for configuring administrating the installation are also included in this document.
...
Convention | Description | Example | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
Bold and Capitalized Command | Indicates a Menu command | Click the Open command. | ||||||||||||
Capitalized command>Capitalized command | Indicates Sequential Menu commands | Admin > Refresh | ||||||||||||
| Used for filenames, directory names, commands, file listings, source code examples and anything that would appear in a Java program, such as methods, variables, and classes. |
| ||||||||||||
Boldface type | Options that you select in dialog boxes or drop-down menus. Buttons or icons that you click. | In the Open dialog box, select the file and click the Open button. | ||||||||||||
Italics | Used to reference other documents, sections, figures, and tables. | caCORE Software Development Kit 1.0 Programmer's Guide | ||||||||||||
Italic boldface type | Text that you type | In the New Subset text box, enter Proprietary Proteins. | ||||||||||||
| Highlights a concept of particular interest |
| ||||||||||||
| Highlights information of which you should be particularly aware. |
| ||||||||||||
{} | Curly brackets are used for replaceable items. | Replace {LEXBIG_DIRECTORY} with its proper value such as |
...
For more information on the LexBIG architecture see Architecture Specification at http://cabigcvs.nci.nih.gov/viewcvs/viewcvs.cgi/lexgrid/LexBIG_architecture_specification.pdf HISTORICAL.
After successfully installing LexBIG and running the verification test suite, as described in this guide, you should be ready to start programming using the API to meet the needs of application needs. If you have the required software installed on your system (see the previous section), then installing and running the test should not take more than 60 minutes.
...
Complete the following steps to download LexBIG:
- To download Download the LexBIG install package go to the NCI GForge web site https://gforge.nci.nih.gov/frs/?group_id=491
- Select the most recent version of the LexBIG Software Package lexbig-install-X.X.X.jar (e.g., 1.0.1, lexbig-install-1.0.1.jar). Save this file to your computer. This location will be referred to as the SAVE_DIRECTORY. You may have to disable Pop-up blockers to allow save the install package to your local computer.
Using Microsoft Windows Environment Command Prompt change directory to the SAVE_DIRECTORY of the LexBIG software package you saved in step 2. At the command prompt enter the following command to begin the installation wizard.
Code Block java -jar lexbig-install-X.X.X.jar
- As an alternative to the command line instruction you can navigate to the SAVE_DIRECTORY with the File Explorer. Double Click on the lexbig-install-1.0.1.jar file. This will launch the install wizard with a typical java installation
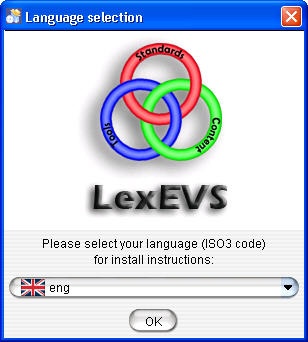
- Select the language and click OK button to begin the installation. Note that the only language currently supported is English.
- As an alternative to the command line instruction you can navigate to the SAVE_DIRECTORY with the File Explorer. Double Click on the lexbig-install-1.0.1.jar file. This will launch the install wizard with a typical java installation
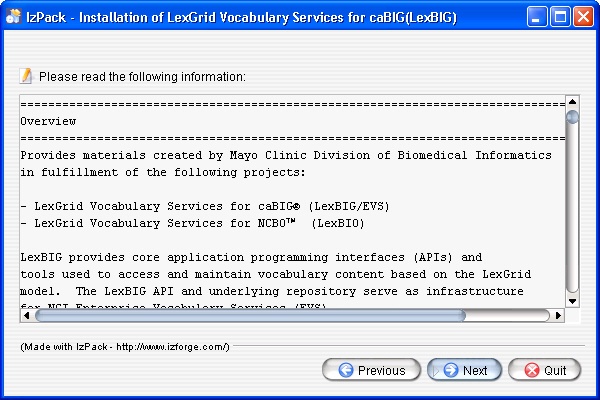
- After the initial welcome screen the release notes for the LexBIG distribution are displayed. Once you have read through the release notes click the Next button to continue.
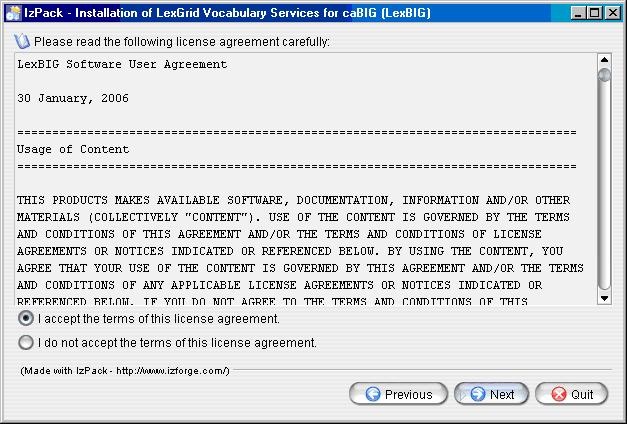
- The next step is to review the license agreement of the LexBIG software and accept the terms of the agreement. Click Next button to continue with installation.
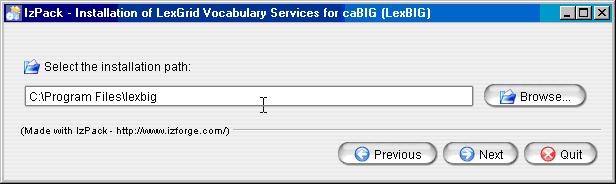
Enter the path where you would like the LexBIG software installed. Click the Next button to continue installation. This will be referred to as the LEXBIG_DIRECTORY throughout the remaining instructions.
Info title Note If the directory does not exist, the program will prompt to proceed with creating the new directory. If the directory does exist, the program will prompt to overwrite the directory and files in the installation path.
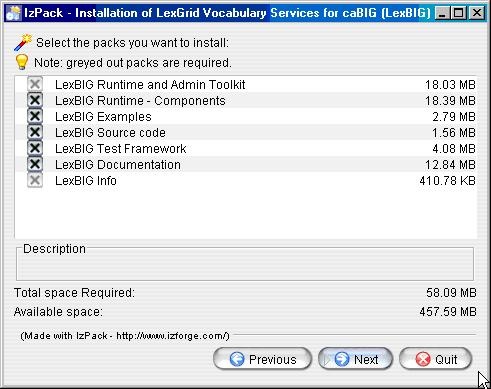
- Select the components to be installed for LexBIG. Two of the components LexBIG Runtime and Admin Toolkit and LexBIG Info are required and cannot be unchecked. The remaining components are optional. Once components have been decided click the Next button to continue with installation.
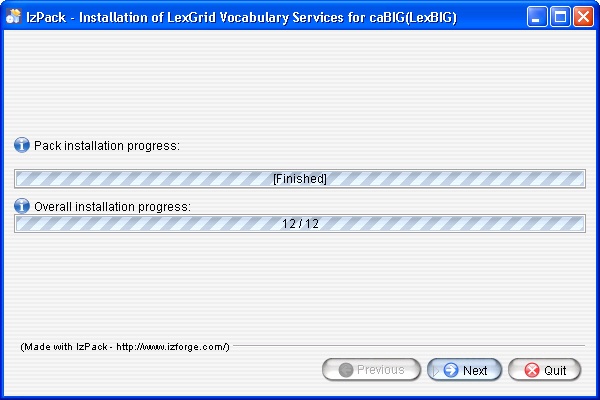
- Once all the components have been installed a Finished prompt will be displayed. Click the Next button to continue installation.
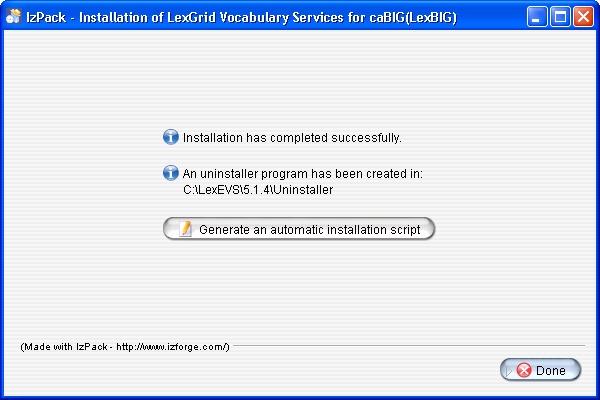
- The last step of the installation wizard provides the ability to generate an automatic installation script that can be used on other machines. This installation script can be used to install LexBIG without graphic wizard. Click Done to complete the installation process.
...
- To download the LexBIG install package go to the NCI GForge web site http://gforge.nci.nih.gov/frs/?group_id=14files archive.
- Select the most recent version of the LexBIG Software Package lexbig-install-X.X.X.jar (e.g., 1.0.1, lexbig-install-1.0.1.jar). Save this file to your computer. This location will be referred to as the SAVE_DIRECTORY. You may have to disable Pop-up blockers to allow save the install package to your local computer.
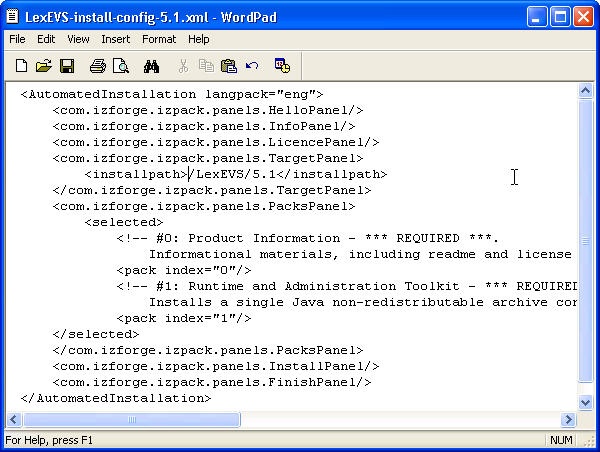
- Select the install-config.xml file. Save this file to your SAVE_DIRECTORY.
- Edit the install-config.xml file to configure the components to be installed. The install path can be modified to the location of choice. Components LexBIG Runtime and Admin Toolkit and LexBIG Info are required. Remove lines in install-config.xml file that you do not want to be installed. By default, the file is configured to install all packages.
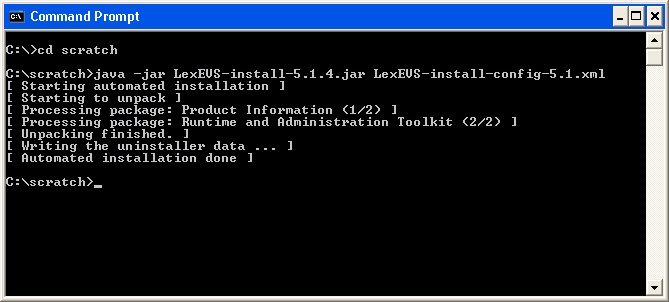
At command prompt in the SAVE_DIRECTORY enter the command:
Code Block java -jar lexbig-install-1.0.1.jar install-config.xml
Configuring the LexBIG Environment
...
| Warning | ||
|---|---|---|
| ||
The LexBIG runtime and database environments must still be configured prior to invoking the test suite, as described above. |
In a Command Promptwindow, enter the following command to go to the test directory:
Code Block cd {LEXBIG_DIRECTORY}/testRun the TestRunner utility to start the test process.
For Windows Environment enter:Code Block TestRunner.bat
For Linux Environment enter:
Code Block TestRunner.sh -h
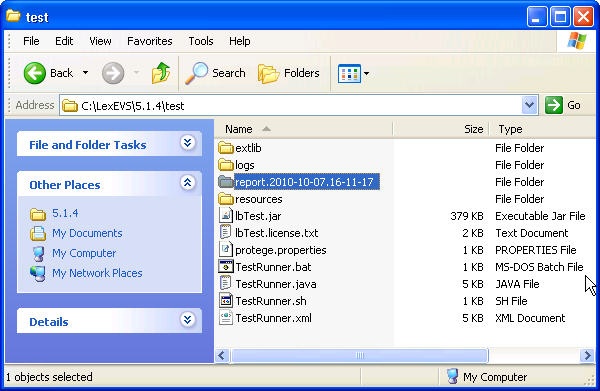
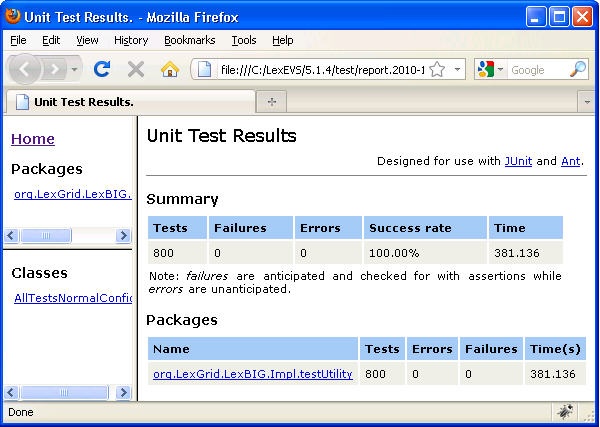
- Use file explorer to navigate to the directory that contains the test report. The report is placed in the {LexBIG_DIRECTORY}/test.
- Navigate to the report that represents the date and time you executed the test.
- Navigate to the report that represents the date and time you executed the test.
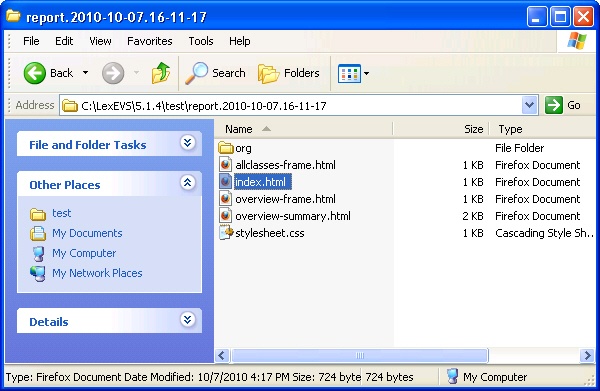
- Review the test results opening index.html file using a web browser.
...
Administrative Program | Description | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
ActivateScheme | Activates a coding scheme based on unique URN and version.
| |||||||||
ClearOrphanedResources | Clean up orphaned resources - databases and indexes.
| |||||||||
DeactivateScheme | Deactivates a coding scheme based on unique URN and version.
| |||||||||
ExportLgXML | Exports content from the repository to a file in the LexGrid canonical XML format.
| |||||||||
ExportOBO | Exports content from the repository to a file in the Open Biomedical Ontologies (OBO) format.
| |||||||||
ExportOWL | Exports content from the repository to a file in OWL format.
| |||||||||
ListExtensions | List registered extensions to the LexBIG runtime environment.
| |||||||||
ListSchemes | List all currently registered vocabularies.
| |||||||||
LoadLgXML | Loads a vocabulary file, provided in LexGrid canonical xml format.
| |||||||||
LoadNCIHistory | Imports NCI History data to the LexBIG repository.
| |||||||||
LoadNCIMeta | Loads the NCI MetaThesaurus, provided as a collection of RRF files.
| |||||||||
LoadNCIThesOWL | Loads an OWL file containing a version of the NCI Thesaurus
| |||||||||
LoadOBO | Loads a file specified in the Open Biomedical Ontologies (OBO) format.
| |||||||||
LoadOWL | Loads an OWL file.
Options:
| |||||||||
LoadUMLSDatabase | Loads UMLS content, provided as a collection of RRF files in a single directory. Files may comprise the entire UMLS distribution or pruned via the MetamorphoSys tool. A complete list of source vocabularies is available online at http://www.nlm.nih.gov/research/umls/metaa1.html.
| |||||||||
LoadUMLSFiles | Loads UMLS content, provided as a collection of RRF files in a single directory. Files may comprise the entire UMLS distribution or pruned via the MetamorphoSys tool. A complete list of source vocabularies is available online at http://www.nlm.nih.gov/research/umls/metaa1.html.
| |||||||||
LoadUMLSSemnet | Loads the UMLS Semantic Network, provided as a collection of files in a single directory. The following files are expected to be provided from the National Library of Medicine (NLM) distribution. These files can be downloaded from the NLM web site at http://semanticnetwork.nlm.nih.gov/Download/index.html.
| |||||||||
LoadFMA | Imports from an FMA database to a LexBIG repository. Requires that the pprj file be configured with a database URN, username, password for an FMA MySQL based database. The FMA.pprj file and MySQL dump file are available at http://sig.biostr.washington.edu/projects/fm/upon registration.
| |||||||||
LoadHL7RIM | Converts an HL7 RIM MS Access database to a LexGrid database
| |||||||||
LoadMetaData | Loads optional XML-based metadata to be associated with an existing coding scheme.
| |||||||||
RebuildIndex | Rebuilds indexes associated with the specified coding scheme.
| |||||||||
RemoveIndex | Clears an optional named index associated with the specified coding scheme. Note: built-in indices required by the LexBIG runtime cannot be removed.
| |||||||||
RemoveScheme | Removes a coding scheme based on unique URN and version.
| |||||||||
TagScheme | Associates a tag ID (e.g. 'PRODUCTION' or 'TEST') with a coding scheme URN and version.
| |||||||||
TestRunner | Located in {LEXBIG_DIRECTORY}/test. Executes a suite of tests for the LexBIG installation.
Options:
| |||||||||
TransferScheme | Tool to help gather information necessary to transfer data from one SQL server to another.
|
...
This LexBIG installation provides the UMLS Semantic Net and a sampling of the NCI Thesaurus content (sample.owl) that can be loaded into the database.
In a Command Promptwindow, enter the following command to go to the example programs.
Code Block cd {LEXBIG_DIRECTORY}/examplesTo load the example vocabularies, run the appropriate LoadSampleData script (LoadSampleData.bat for Windows; LoadSampleData.sh for Linux).
Info title Note Vocabularies should not be loaded until configuration of the LexBIG runtime and database server are complete.
Running the Sample Query Programs
A set of sample programs are provided in the {LEXBIG_DIRECTORY}/examples directory. To run the sample query programs successfully a vocabulary must have been loaded.
Enter the following command:
Code Block cd {LEXBIG_DIRECTORY}/examplesExecute one of sample programs. (.bat for windows or .sh for Linux.)
Code Block FindConceptNameForCode.bat
Code Block FindPropsandAssocForCode.bat
Code Block FindRelatedCodes
Code Block FindTreeforCodeAndAssoc
...
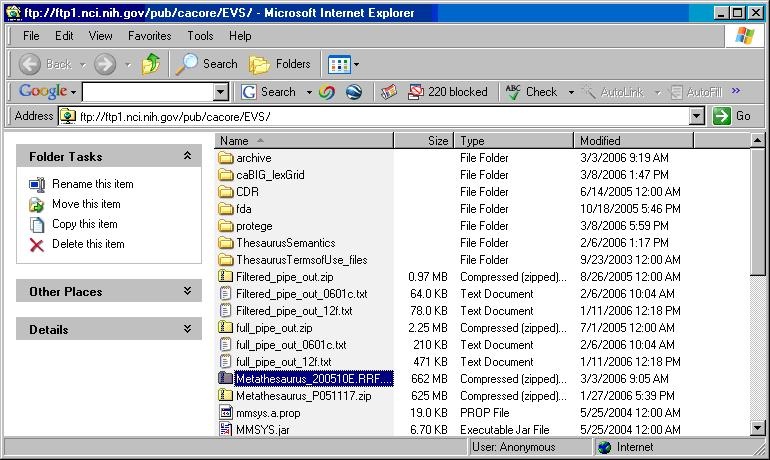
- Using a web or ftp client go to URL: ftp://ftp1.nci.nih.gov/pub/cacore/EVS/
- Select the version of NCI Thesaurus OWL you wish to download and save the file to a directory on your machine.
- Extract the OWL file from the zip download and save in a directory on your machine. This directory will be referred to as NCI_THESAURUS_DIRECTORY
Using the LexBIG utilities load the NCI Thesaurus:
Code Block cd {LexBIG_DIRECTORY}/adminFor Windows installation use the following command
Code Block LoadNCIThesOWL.bat -nf -in "file:///{NCI_THESAURUS_DIRECTORY}/Thesaurus_05.12f.owl"For Linux installation use the following command
Code Block LoadNCIThesOWL.sh -nf -in "file:///{NCI_THESAURUS_DIRECTORY}/Thesaurus_05.12f.owl"
...
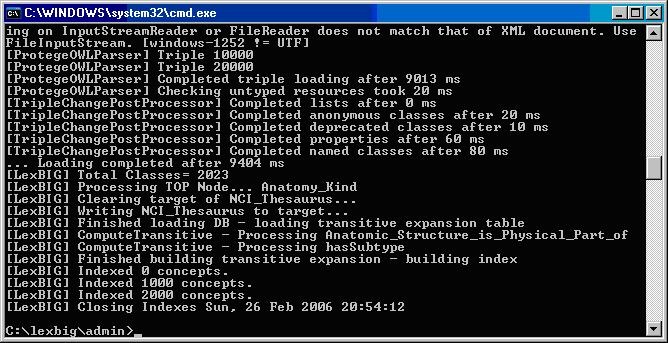
The following example shows output from load of NCI Thesaurus 05.12f.
| Code Block |
|---|
...
[LexBIG] Processing TOP Node... Retired_Kind
[LexBIG] Clearing target of NCI_Thesaurus...
[LexBIG] Writing NCI_Thesaurus to target...
[LexBIG] Finished loading DB - loading transitive expansion table
[LexBIG] ComputeTransitive - Processing Anatomic_Structure_Has_Location
[LexBIG] ComputeTransitive - Processing Anatomic_Structure_is_Physical_Part_of
[LexBIG] ComputeTransitive - Processing Biological_Process_Has_Initiator_Process
[LexBIG] ComputeTransitive - Processing Biological_Process_Has_Result_Biological_Process
[LexBIG] ComputeTransitive - Processing Biological_Process_Is_Part_of_Process
[LexBIG] ComputeTransitive - Processing Conceptual_Part_Of
[LexBIG] ComputeTransitive - Processing Disease_Excludes_Finding
[LexBIG] ComputeTransitive - Processing Disease_Has_Associated_Disease
[LexBIG] ComputeTransitive - Processing Disease_Has_Finding
[LexBIG] ComputeTransitive - Processing Disease_May_Have_Associated_Disease
[LexBIG] ComputeTransitive - Processing Disease_May_Have_Finding
[LexBIG] ComputeTransitive - Processing Gene_Product_Has_Biochemical_Function
[LexBIG] ComputeTransitive - Processing Gene_Product_Has_Chemical_Classification
[LexBIG] ComputeTransitive - Processing Gene_Product_is_Physical_Part_of
[LexBIG] ComputeTransitive - Processing hasSubtype
[LexBIG] Finished building transitive expansion - building index
[LexBIG] Getting a results from sql (a page if using mysql)
[LexBIG] Indexed 0 concepts.
[LexBIG] Indexed 5000 concepts.
[LexBIG] Indexed 10000 concepts.
[LexBIG] Indexed 15000 concepts.
[LexBIG] Indexed 20000 concepts.
[LexBIG] Indexed 25000 concepts.
[LexBIG] Indexed 30000 concepts.
[LexBIG] Indexed 35000 concepts.
[LexBIG] Indexed 40000 concepts.
[LexBIG] Indexed 45000 concepts.
[LexBIG] Indexed 46000 concepts.
[LexBIG] Getting a results from sql (a page if using mysql)
[LexBIG] Closing Indexes Mon, 27 Feb 2006 01:44:22
[LexBIG] Finished indexing
|
...
- Using a web or ftp client go to URL: ftp://ftp1.nci.nih.gov/pub/cacore/EVS/
- Select the version of NCI Metathesaurus RRF you wish to download and save the file to a directory on your machine.
Extract the RRF files from the zip download and save in a directory on your machine. This directory will be referred to as NCI_METATHESAURUS_DIRECTORY.
Info title Note RELASE_INFO.RRF is required to be present for the load utility to work.
Using the LexBIG utilities load the NCI Thesaurus
Code Block cd {LexBIG_DIRECTORY}/adminFor Windows installation use the following command
Code Block LoadNCIMeta.bat -nf -in "file:///{NCI_METATHESAURUS_DIRECTORY}/"For Linux installation use the following command
Code Block LoadNCIMeta.sh -nf -in "file:///{NCI_THESAURUS_DIRECTORY}/"
...
- Using a web or ftp client go to URL: ftp://ftp1.nci.nih.gov/pub/cacore/EVS/
<TO BE DETERMINED> - Select the version of NCI History you wish to download. Save the file to a directory on your machine. Select the VersionFile download to the same directory as the history file.
- Extract the History files from the zip download and save in a directory on your machine. This directory will be referred to as NCI_HISTORY_DIRECTORY
Using the LexBIG utilities load the NCI Thesaurus
Code Block cd {LexBIG_DIRECTORY}/adminFor Windows installation use the following command
Code Block LoadNCIHistory.bat -nf -in "file:///{NCI_HISTORY_DIRECTORY}" -vf "file:///NCI_HISTORY_DIRECTORY}/VersionFile"For Linux installation use the following command
Code Block LoadNCIHistory.sh -nf -in "file:///{NCI_HISTORY_DIRECTORY}" -vf "file:///NCI_HISTORY_DIRECTORY}/VersionFile"
...
This section describes the steps to deactivate a coding scheme and remove coding scheme from LexBIG Service.
Change directory to LexBIG administration directory. Enter:
Code Block cd {LEXBIG_DIRECTORY}/adminUse the DeactiveScheme utility to prevent access to coding scheme. Once a coding scheme is deactivated, client programs will not be able to access the content for the specific coding scheme and version. Example:
Code Block DeactivateScheme -u "urn:oid:2.16.840.1.113883.3.26.1.1" -v "05.12f"
Use RemoveScheme utility to remove coding scheme from LexBIG service and database. Example:
Code Block RemoveScheme -u "urn:oid:2.16.840.1.113883.3.26.1.1" -v "05.12f"
...
This section describes the steps to tag a coding scheme to be used via LexBIG API.
Change directory to LexBIG administration directory. Enter:
Code Block cd {LEXBIG_DIRECTORY}/adminUse the TagScheme to tag a coding system and version with a local tag name (e.g. PRODUCTION). This tag name can be used via LexBIG API for query restriction. Example:
Code Block TagScheme -u "urn:oid:2.16.840.1.113883.3.26.1.1" -v "05.12f" -t "PRODUCTION"
...
The registered name is the key used to find a coding scheme (for example a unique URL or namespace by which other people access same coding scheme). This String value will be used to identify the manifest entry in the manifest file for the coding scheme too. For example the registered name for coding scheme "Amino-acid" is http://www.co-ode.org/ontologies/amino-acid/2006/05/18/amino-acid.owl#. This string is also set as the coding scheme's registered name field in the LexGrid model.
...
An example code snippet:
| Code Block |
|---|
LexBIGService lbs = new LexBIGServiceImpl();
LexBIGServiceManager lbsm = lbs.getServiceManager(null);
OWL_Loader loader = (OWL_Loader) lbsm.getLoader("OWLLoader");
if (toValidateOnly)
\{
loader.validate(source, manifest, vl);
System.out.println("VALIDATION SUCCESSFUL");
\}
else
\{
loader.load(new File("resources/testData/amino-
cid.owl").toURI(),
new File("resources/testData/aa-manifest.xml").toURI(),true,
true);
\}
|
For all other manifest loads the following methods are employed.
| Code Block |
|---|
// Find the registered extension handling this type of load
LexBIGService lbs = new LexBIGServiceImpl();
LexBIGServiceManager lbsm = lbs.getServiceManager(null);
HL7_Loader loader = (HL7_Loader)lbsm.getLoader(org.LexGrid.LexBIG.Impl.loaders.HL7LoaderImpl.name);
// updated to include manifest
loader.setCodingSchemeManifestURI(manifest);
// updated to include loader preferences
loader.setLoaderPreferences(loaderPrefs);
loader.load(dbPath, stopOnErrors, true);
|
...
Loader preferences for a given loader can be loaded from a file in the following format example for the NCI OWL loader.
| Code Block |
|---|
<?xml version="1.0" encoding="UTF-8"?>
<OWLLoaderPreferences xmlns:basePreferences="http://LexGrid.org/LexBIG/LoadPreferences">
<MatchConceptCode>currency</MatchConceptCode>
<PrioritizedCommentNames>
<name>units</name>
</PrioritizedCommentNames>
<PrioritizedDefinitionNames>
<name>max</name>
</PrioritizedDefinitionNames>
<PrioritizedPresentationNames>
<name>currency</name>
</PrioritizedPresentationNames>
</OWLLoaderPreferences>
|
...
LexBIG Application Support can be contacted at:
Division of Biomedical Statistics and Informatics
Mayo Clinic
200 1st ST SW
Rochester, MN 55905
informatics@mayo.edu
Discussion forums, trackers for feature and bug submissions, and news are maintained at NCI Gforge for LexBIG: https://gforge.nci.nih.gov/projects/lexevs/
...
| Scrollbar | ||
|---|---|---|
|