|
Page History
| Scrollbar | ||
|---|---|---|
|
Problem: When uploading data from large microarrays, the size of your data archive may exceed the individual file size limit of 2 GB.
Topic: caArray Usage
Release: caArray 2.0 and above
Date entered: 10/17/2011
Solution
This article presents a workaround which allows you to break down your dataset into smaller, more manageable chunks that can be individually uploaded without violating the 2 GB limit.
Overview
Your experiment dataset consists of an IDF metadata file and its corresponding SDRF metadata file, which, in turn, is associated with one or more raw and derived array data files. In this tutorial, the array files we will use are in the Agilent TXT (raw) and TSV (derived) formats; the file formats for your data may differ.
...
- Divide the array data files into smaller batches, each of which will be no larger than 2 GB following ZIP compression.
- Split the original SDRF file into multiple SDRF files, each corresponding to a single batch and referencing only the array data files from that batch.
- Create multiple IDF files derived from the original IDF, with each one uniquely referencing one of the SDRF files created in the previous step.
- Create a ZIP archive for each batch, containing a single IDF and its associated SDRF and raw and array data files.
- Upload each ZIP archive individually, then validate and import the files from each.
Prerequisites
This tutorial assumes that you have past experience and basic familiarity with uploading data into caArray. Specifically, it assumes that you have already created an experiment for your data, uploaded the corresponding array design, and associated the experiment with that design. In case you lack a basic background on uploading caArray data, please refer to the official caArray User's Guide on the NCI wiki at https://wiki.nci.nih.gov/x/LBo9Ag.
You must have all your experiment data readily accessible on your computer (i.e., not archived or compressed). The data should preferably be consolidated into a single location (i.e., a folder containing every single IDF, SDRF, raw and derived array data file from the experiment). You will also need an archive creation utility installed on your computer. In this tutorial, we will use WinZip (www.winzip.com), but any comparable utility with support for the ZIP format will do.
Reference Information
The experiment data used in this tutorial was not generated de novo; it came from an existing experiment whose data is publicly available on the official NCI instance of caArray at https://array.nci.nih.gov/caarray/home.action (note that you may download this data without registering for an account on the site). The experiment, entitled "TCGA Ovarian: Comparative Genome Hybridization Analysis Using the Agilent Human Genome CGH 244A Platform", was conducted at Harvard Medical School in Boston, MA. It can be accessed via the URL https://array.nci.nih.gov/caarray/project/EXP-498 or by searching for the experiment ID 'EXP-498' on the NCI caArray instance. The array design used was TCGA-Agilent_HG-CGH-244A; the array design files can be downloaded from the experiment in ADF format, as can all the experiment data, including the IDF and SDRF metadata files, the Agilent TXT raw array data files, and the TSV derived array data files.
Getting Started -- Dividing the Array Data Into Batches
The screenshot below shows a portion of the dataset from our sample experiment, including the IDF and SDRF files, as well as some TXT and TSV files.
...
- Create a separate subfolder for each new batch
- Select multiple data files in your file manager, taking care to keep selection size below 5 GB uncompressed (2 GB compressed)
- Move selected files to respective batch folder
Splitting The Original SDRF File
Now that we've created batches of our array data files, our next step is to split the original SDRF file into multiple SDRFs, each corresponding to a single batch and referencing only the array data files from that batch. To do so, first open the original SDRF file in Microsoft Excel or another tab-limited data viewer, as shown below:
...
- Open the original SDRF file and locate the rows referencing the array files for the next batch
- Delete all other rows except for the top header row
- Save the modified SDRF as a new file with a filename unique to its respective batch
- Copy the newly generated SDRF to its respective batch's folder
Creating a Unique IDF File For Each Batch
Once you've generated a unique SDRF file for each batch, you must also generate a unique IDF file which references that SDRF file. You can do so simply by opening the original IDF file and editing the field 'SDRF Files' with the filename of the SDRF you wish to reference, as shown below:
...
- Open the original IDF file and locate the 'SDRF Files' field.
- Edit this field to reflect the file name of the SDRF file you wish to reference.
- Save the modified IDF file with a unique filename that is parallel to the referenced SDRF's filename.
- Copy the newly generated IDF to its respective batch's folder
Creating the Archives
Now that we've divided our dataset into batches and generated the corresponding IDF and SDRF files for each, our next step is to create a ZIP archive of each batch. Launch WinZip, click the 'New' toolbar button, and enter a name for your archive in the 'New Archive' dialog. We'll call ours 'upload.zip', as shown below.
...
In our example, the 'upload.zip' data archive we created is approximately 900 MB in size, which is below the 2 GB upload limit. If your data archive turns out to be larger than 2 GB, you will not be able to upload it until you re-create it with a higher compression ratio.
Uploading the Archive
To upload the archive, first log in to caArray and navigate to the experiment you will be upload your data into, then select the 'Data' tab, followed by the 'Manage Data' tab beneath it. Now click on the 'Upload New Files' button as shown below.
...
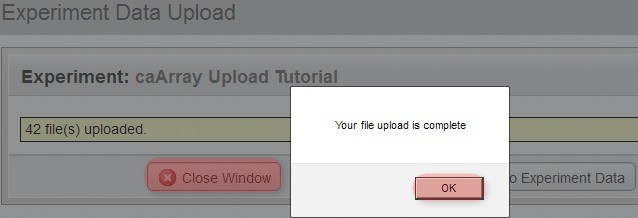
You'll know when the upload is complete when you see a new window overlaid over the upload window with the message, 'Your file upload is complete'.
Validating the Archive
Back in the main experiment window, the contents of the archive we just uploaded are now listed under the 'Manage Data' tab. The TSV matrix files are considered supplemental, so we will move them to the 'Supplemental Files' tab by first using the 'Filter By File Type' drop-down to show only TSV files, then checking off all the TSV files in the list, and finally clicking on the 'Add Supplemental Files' button below.
...
The imported files now appear under the 'Imported Data' tab with a status of 'Imported' alongside other files from a previous upload to the same experiment.
Reproducing the Procedure
So far, only one-sixth of the data has been uploaded. You can reproduce the procedure we followed so far to upload the data from your experiment. The procedure, summarized below, is as follows:
...
- Import the validated files into the experiment
Have a comment?
Please leave your comment in the caArray End User Forum.
...