To Print the Guide
We recommend you print one wiki page of the guide at a time. To do this, click the printer icon at the top right of the page; then from the browser File menu, choose Print. Printing multiple pages at one time is more complex. For instructions, refer to How do I print multiple pages?.
Searching for Compounds Associated with a Disease
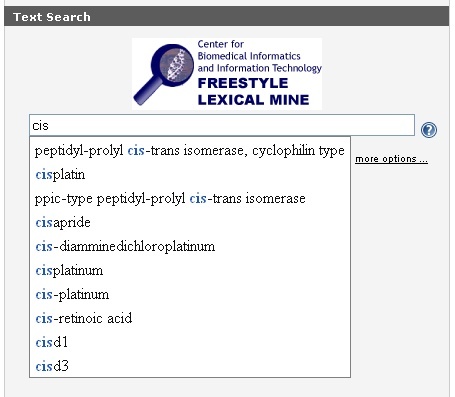
Because the Freestyle Lexical Mine suggests caBIO terms that match the characters you have entered, it is relatively easy to find a compound search term that matches your desired disease concept.
If you would like to search for any disease term that contains a string (set of alphanumeric characters) use the special character "*" which will retrieve results will have zero or more characters in place of the asterisk. For example, "cisplatin" would only return objects that are associated with this exact compound term, but "cis*" will retrieve objects that are associated with any term that contains the string "cis."
After entering a search term (1), click the Submit button (2) to retrieve results. Although you may limit your search by clicking on the more options ... link, this is not required (3).
Tip
If you have cannot find an appropriate search term, click the Contact Us link for help.
Search Results
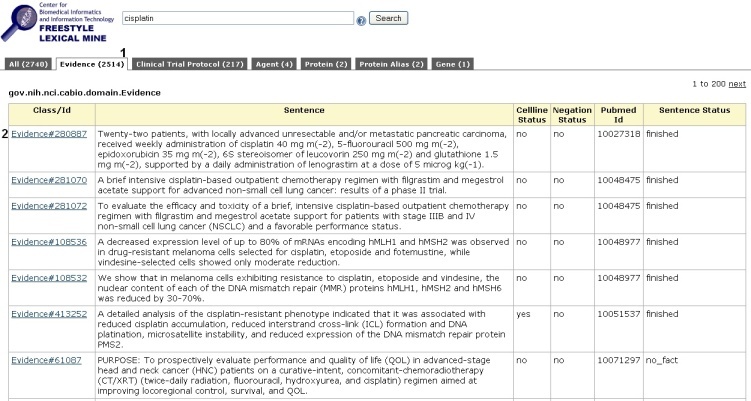
The caBIO Portal Freestyle Lexical Mine will retrieve objects that match your search term. These objects are grouped by type, which are shown as tabs at the top of the results page. To view genes that are associated with your compound term, click the Evidence tab at the top of the page (1).
Each row in the Evidence results table is a truncated view of an Evidence object (i.e., not all attributes and methods are shown on this page), whereas the columns show the attributes associated with the Evidence type: class and identifier (Class/Id), the evidence of the gene-disease association (Sentence), whether the evidence was collected from experiments involving cell lines (Cellline Status), whether the evidence is negative (i.e., gene X is not associated with disease Y; Negation Status), the PubMed identifier for the abstract from which the evidence was extracted (Pubmed Id), and whether the status of the sentence (Sentence Status). For additional information on the data, metadata, and annotations of the Evidence type objects, refer to the section Data, Metadata, and Annotations.
Note
Although the Cancer Gene Index refers to pharmacological substances as "compounds," caBIO and the NCI Thesaurus use the term "agent" for the same concept.
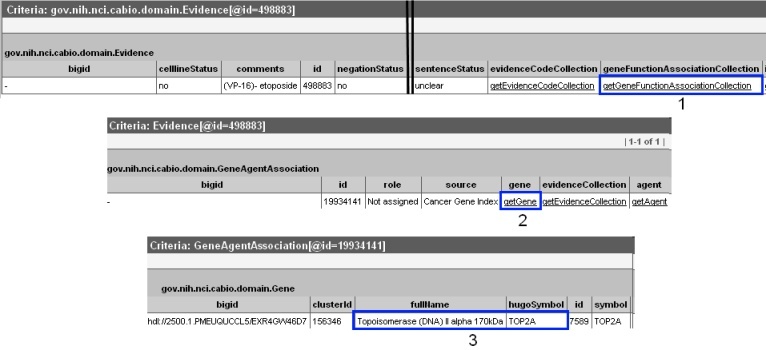
To discover which gene is associated with each piece of evidence, click on the Class/Id link for the desired object. This will open the full Evidence type object. Scroll over to the right, and click on getGeneFunctionAssociationCollection method link (1) to view the Gene Agent Association type object. This object has a role attribute that contains one or more https://wiki.nci.nih.gov/x/zC1yAQ#Role Codes or https://wiki.nci.nih.gov/x/zC1yAQ#Role Details that describe the nature of the gene-compound/agent relationship, as well as a notation that the Cancer Gene Index is the source of these data.
Warning
If you do not want to spend time navigating through the caBIO object model for candidate gene-compound/agent associations that were found to be false positives, unclear, or redundant to other data, select only Evidence objects where the Sentence Status is finished.
Note
A single piece of evidence may have multiple Role Codes and Role Details describing the gene-compound/agent association, and the evidence may also describe gene-disease associations. Thus, after clicking the getGeneFunctionAssociationCollection link, you may see multiple retrieved objects of type gov.nih.nci.cabio.domain.GeneAgentAssociation and even multiple object retrieved records of type gov.nih.nci.cabio.domain.GeneDiseaseAssociation.
Click on the getGene link (2) to access the related Gene object (bottom panel). This Gene object contains the full name and HUGO Gene Symbol in the fullName and hugoSymbol columns (3), for example, for the gene associated with the disease of interest and a specific piece of evidence.
To explore additional genes associated with the compound term of interest, navigate back to the evidence page and repeat this process.
Be Careful
If you find yourself in a part of the object model that you do not understand or if you get confused, stop and navigate your web browser back to the search results page with the Evidence tab. For your reference, the subset of caBIO classes that are related to the Cancer Gene Index are shown in the figure below.
The full model is available on the caBIO gForge page.
Agent Ontologies
If you would like to search for genes that are associated with parent, sister, or child concepts to your compound/agent search term, you must use the NCI Thesaurus.