Contents of this Page
caMicroscope Docker Distribution
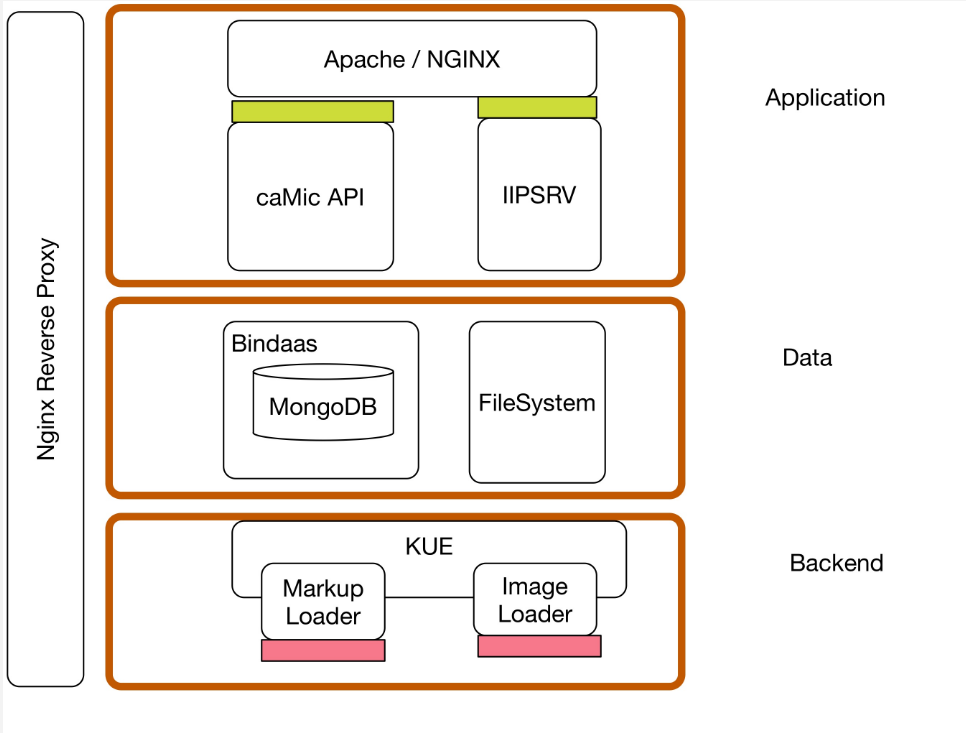
Architecture
- Application => Viewer
- Data => Data
- Backend => Loader
Installation
Build images
docker build -t camicroscope_data data/docker build -t camicroscope_loaders loaders/docker build -t camicroscope_viewer viewer/
These builds take about an hour, so feel free to grab a coffee.
Running the containers
Data
- Set environment variables on host machine for Images and Data directories
export CAMIC_IMAGES_DIR=<path to images directory>export CAMIC_DATA_DIR=<path to data directory> - Set Bindaas port
$CAMIC_BINDAAS_PORT=9099docker run -itd -p $CAMIC_BINDAAS_PORT:9099 -v $CAMIC_IMAGES_DIR:/data/images -v $CAMIC_DATA_DIR:/data/db camicroscope_data - Run
docker inspectto find the IP address for Data container. - Use
<CAMIC_DATA_IP>as a placeholder.
Loaders
- Set up environment variables:
$CAMIC_KUE_PORT=5000
$CAMIC_MARKUPLOADER_PORT=6000$CAMIC_DATALOADER_PORT=6002 - Run the container:
docker run -itd -p $CAMIC_KUE_PORT:3000 -p $CAMIC_MARKUPLOADER_PORT:3001 -p $CAMIC_DATALOADER_PORT:3002 -v $CAMIC_IMAGES_DIR:/data/images -e "dataloader_host=http://<CAMIC_DATA_IP>:9099" -e "annotations_host=http://<CAMIC_DATA_IP>:9099" camicroscope_loaders
Note that -p $CAMIC_KUE_PORT:3000 -p $CAMIC_MARKUPLOADER_PORT:3001 -p $CAMIC_DATALOADER_PORT:3002 is optional.
Viewer
- Set up environment variables
:
$CAMIC_VIEWER_PORT=1337
<HTML_DIRECTORY> - Run:
docker run -itd -p $CAMIC_VIEWER_PORT:80 -v <HTML_DIRECTORY>/html:/var/www/html -v $CAMIC_IMAGES_DIR:/data/images camicroscope_viewer
You should now be able to load the images and see them at http://localhost:$CAMIC_VIEWER_PORT/camicroscope2/osdCamicroscope.php?tissueId=<Case_ID>