Most OWL formatted loads make use of the least complicated procedure to achieve an installation. However, it is a popular format and LexEVS offers some extensive options for customizing a terminology at load time. As such, we'll go through an OWL load step by step.
1 |
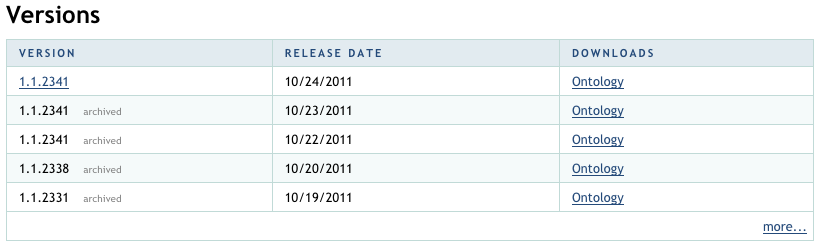
Obtain an OWL formatted source. |
2 |
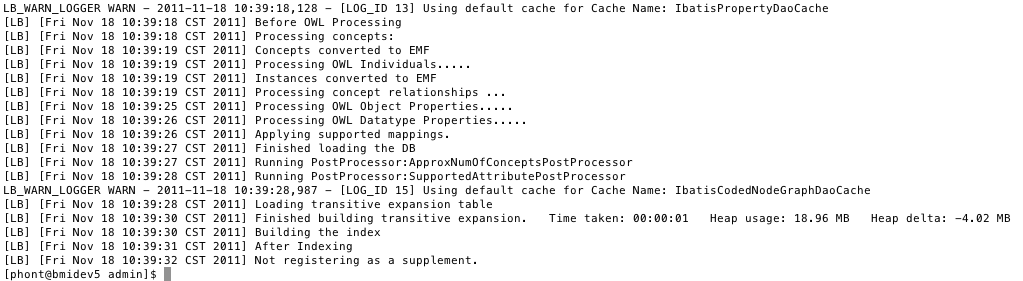
Load the source without options. ./LoadOWL.sh -in "file:///home/LargeStorage/ontologies/owl/amino-acid.owl" Load in Windows environments: LoadOWL.bat -in "file:///home/LargeStorage/ontologies/owl/amino-acid.owl" |
3 |
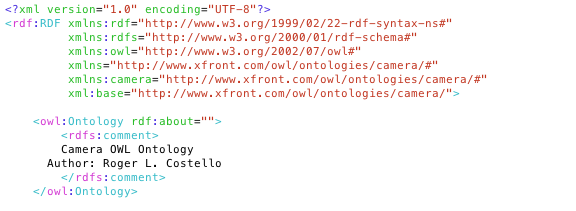
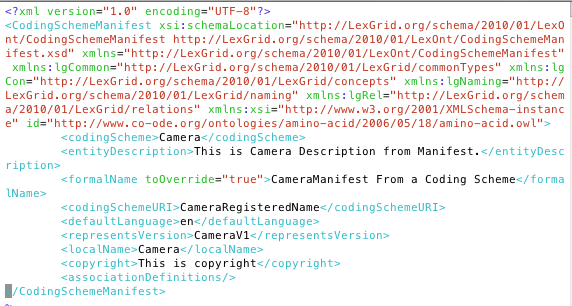
Load source with options: ./LoadOWL.sh -in "file:///data/phont/LexEVS/test/resources/testData/camera.owl" -mf "file:///data/phont/LexEVS/test/resources/testData/Camera-manifest.xml" Loading in Windows LoadOWL.bat -in "file:///data/phont/LexEVS/test/resources/testData/camera.owl" -mf "file:///data/phont/LexEVS/test/resources/testData/Camera-manifest.xml"
|