- Vocabulary Administration Overview
- Lucene Index Administration
- Misc Data Base Administration Scripts
- LexEVS Export Scripts
- LexEVS Loader Scripts
- Special Batch Loading Functions
- Scheme and Metadata Removal
- Pick List and Value Set Load Administration
- LexEVS Validation Test Script
- Displaying Current Coding Schemes
- Using optional parameters
- Setting Java Virtual Machine Options
- Running the sample query programs
- NCI Thesaurus vocabulary
- NCI Metathesaurus vocabulary
- NCI History
- Deactivating and Removing a Vocabulary
- Tagging a Vocabulary
- IndexManagement
- Value Set and Pick List Loading
Vocabulary Administration Overview
A set of administrative shell scripts are provided to manage the LexEVS Service. These scripts are provided for Windows (.bat) and Linux (.sh) operating systems and call wrapper classes for LexEVS administration API classes. These scripts are located in the {LEXBIG_DIRECTORY}/admin and {LEXBIG_DIRECTORY}/test directory. A full description of the options with an example is provided for each command line script.
If you are loading a large vocabulary such as SNOMED or the NCI Metathesaurus, then we recommend the use of these scripts over the GUI since adjusting system memory requirements can be accomplished by changing the load script with a text editor.
Coding Scheme Metadata Access and Administration
Shell Script |
Use and Function |
|---|---|
ActivateScheme |
Activates a coding scheme based on unique URN and version.Options: -u,-urn <urn> URN uniquely identifying the code system. -v,-version <versionId> Version identifier. -f,-force Force activation (no confirmation). |
DeactivateScheme |
Deactivates a coding scheme based on unique URN and version.Options: -u,-urn <urn> URN uniquely identifying the code system. -v,version <versionId> Version identifier. -d,date <yyyy-MM-dd,HH:mm:ss> Date and time for deactivation to take effect; immediate if not specified. -f,-force Force deactivation (no confirmation). |
ListExtensions |
List registered extensions to the LexEVS runtime environment.Options: -a,-all List all extensions (default, override by specifying other options). -i,index List index extensions. -m,match List match algorithm extensions. -s,sort List sort algorithm extensions. -g,-generic List generic extensions. |
ListSchemes |
List all currently registered vocabularies.Options: -b,-brief List only coding scheme name, version, urn, and tags (default). -f,-full List full detail for each scheme. |
TagScheme |
Associates a tag ID (e.g. 'PRODUCTION' or 'TEST') with a coding scheme URN and version.Options: -u,-urn <urn> URN uniquely identifying the code system. -v,version <versionId> Version identifier. -t,-tag The tag ID (e.g. 'PRODUCTION' or 'TEST') to assign. |
Shell Script |
Use and Function |
|---|---|
<center>ActivateScheme</center> |
Activates a coding scheme based on unique URN and version. |
Options:
u,-urn <urn> URN uniquely identifying the code system.
v,-version <versionId> Version identifier.
f,-force Force activation (no confirmation).
Example:
ActivateScheme -u "urn:oid:2.16.840.1.113883.3.26.1.1" -v "05.09e" |
<center>DeactivateScheme</center> |
Deactivates a coding scheme based on unique URN and version. |
Options:
u,-urn <urn> URN uniquely identifying the code system.
v,-version <versionId> Version identifier.
d,-date <yyyy-MM-dd,HH:mm:ss> Date and time for deactivation to take effect; immediate if not specified.
f,-force Force deactivation (no confirmation).
Example:
DeactivateScheme -u "urn:oid:2.16.840.1.113883.3.26.1.1" -v "05.09e" -d "01/31/2099,12:00:00 |
<center>ListExtensions</center> |
List registered extensions to the LexEVS runtime environment. |
Options:
a,-all List all extensions (default, override by specifying other options).
i,-index List index extensions.
m,-match List match algorithm extensions.
s,-sort List sort algorithm extensions.
g,-generic List generic extensions.
Example:
ListExtensions --a |
<center>ListSchemes</center> |
List all currently registered vocabularies. |
Example:
ListSchemes |
<center>TagScheme</center> |
Associates a tag ID (e.g. 'PRODUCTION' or 'TEST') with a coding scheme URN and version. |
Options:
u,-urn <urn> URN uniquely identifying the code system.
v,-version <versionId> Version identifier.
t,-tag The tag ID (e.g. 'PRODUCTION' or 'TEST') to assign.
Example:
TagScheme -u "urn:oid:2.16.840.1.113883.3.26.1.1" -v 05.09e" -t "TEST" |
Lucene Index Administration
Shell Script |
Use and Function |
|---|---|
<center>CleanUpLuceneIndex</center> |
Clean up orphaned indexes. |
Options:
-h, --help, Prints usage information
r,-reindex, Reindex any missing indicies.
Note: Lucene Clean Up can only be executed in the default Single Index Mode.
Example:
CleanUPLuceneIndex -r |
<center>OptimizeLuceneIndex</center> |
Options: |
Optimizes the Common Lucene Index. |
<center>RebuildIndex</center> |
Rebuilds indexes associated with the specified coding scheme. |
Options:
u,-urn <urn> URN uniquely identifying the code system.
v,-version <versionId> Version identifier.
i,-index <name> Name of the index extension to rebuild (if absent, rebuilds all built-in indices and named extensions).
f,-force Force clear (no confirmation).
Example:
RebuildIndex -u "urn:oid:2.16.840.1.113883.3.26.1.1" -v "05.09e" -i "myindex" |
<center>RemoveIndex</center> |
Clears an optional named index associated with the specified coding scheme. Note: built-in indices required by the LexEVS runtime cannot be removed. |
Options
u,-urn <urn> URN uniquely identifying the code system.
v,-version <versionId> Version identifier.
i,-index <name> Name of the index extension to clear.
f,-force Force clear (no confirmation).
Example: RemoveIndex -u "urn:oid:2.16.840.1.113883.3.26.1.1" -v "05.09e" |
Misc Data Base Administration Scripts
Shell Script |
Use and Function |
|---|---|
<center>ExportDDLScripts</center> |
|
<center>PasswordEncryptor</center> |
Encrypts the given password. |
Argument:
p,-password password for encryption.
Example: PasswordEncryptor -p lexgrid |
LexEVS Export Scripts
Shell Script |
Use and Function |
|---|---|
<center>ExportLgXML</center> |
Exports content from the repository to a file in the LexGrid canonical XML format. |
usage: ExportLgXML [VKC:-xc] [VKC:-an null] [VKC:-xall] -v null -u name -out uri [VKC:-xa] [VKC:-f]
an,-associationsName Export associations with this name. Only valid with export type 'xa'
f,-force If specified, allows the destination file to be overwritten if present.
out,-output <uri> URI or path of the directory to contain the resulting XML file. The file name will be automatically derived from the coding scheme name.
u,-urn <name> URN or local name of the coding scheme to export.
v,-version The assigned tag/label or absolute version identifier of the coding scheme.
xa,-exportAssociationsType of export: export only associations.
xall,-exportAll Type of export: export all content. Default behavior.
xc,-exportConcepts Type of export: export only concepts.
Example:
ExportLgXML -out "file:///path/to/dir" -u "NCI Thesaurus" -v "05.06e" -f |
<center>ExportOBO</center> |
Exports content from the repository to a file in the Open Biomedical Ontologies (OBO) format. |
Options:
out,-output <uri> URI or path of the directory to contain the resulting OBO file. The file name will be automatically derived from the coding scheme name.
u,-urn <name> URN or local name of the coding scheme to export.
v,-version <id> The assigned tag/label or absolute version identifier of the coding scheme.
nf,-noFail If specified, indicates that processing should not stop for recoverable errors
f,-force If specified, allows the destination file to be overwritten if present.
Note: If the coding scheme and version values are unspecified, a list of available coding schemes will be presented for user selection.
Example: ExportOBO -out "file:///path/to/dir" -nf -f
Example: ExportOBO -out "file:///path/to/dir" -u "FBbt" -v "PRODUCTION" -nf -f |
<center>ExportOwlRdf</center> |
Exports content from the repository to a file in OWL format. |
Options:
out,-output <uri> URI or path of the directory to contain the resulting OWL file. The file name will be automatically derived from the coding scheme name.
u,-urn <name> URN or local name of the coding scheme to export.
v,-version <id> The assigned tag/label or absolute version identifier of the coding scheme.
f,-force If specified, allows the destination file to be overwritten if present.
Note: If the URN and version values are unspecified, a list of available coding schemes will be presented for user selection.
Example: ExportOwlRdf -out "file:///path/to/dir" -f
Example: ExportOwlRdf -out "file:///path/to/dir" -u "sample" -v "1.0" -f |
LexEVS Loader Scripts
Shell Script |
Use and Function |
|---|---|
<center>LoadLgXML</center> |
Loads a vocabulary file, provided in LexGrid canonical xml format. |
Options:
in,-input <uri> URI specifying location of the source file.
-v, --validate <level> Perform validation of the candidate resource without loading data. If specified, the '-nf', -a' and '-t' options are ignored. Supported levels of validation include:
0 = Verify document is well-formed
1 = Verify document is valid
nf,-noFail If specified, indicates that processing should not stop for recoverable errors.
-a, --activate ActivateScheme on successful load; if unspecified the vocabulary is loaded but not activated.
-t, --tag <tagID> An optional tag ID (e.g. 'PRODUCTION' or 'TEST') to assign.
Load Example:
LoadLgXML -in "file:///path/to/file.xml" -nf --a
Validation Example:
LoadLgXML -in "file:///path/to/file.xml" -v 0 |
<center>LoadNCIHistory</center> |
Imports NCI History data to the LexEVS repository. |
Options:
in,-input <uri> URI specifying location of the history file
vf,-versionFile <uri> URI specifying location of the file containing version identifiers for the history to be loaded.
-v, --validate <level> Perform validation of the candidate resource without loading data. If specified, the '-nf' and '-r' options are ignored. Supported levels of validation include:
0 = Verify top 10 lines are correct format
1 = Verify correct format for the entire file
nf,-noFail If specified, indicates that processing should not stop for recoverable errors
-r, --replace If not specified, the provided history file will be added into the current history database; otherwise the current database will be replaced by the new content.
Load Example:
LoadNCIHistory --nf -in "file:///path/to/history.file" --vf "file:///path/to/version.file![]() "
"
Validation Example:
LoadNCIHistory -in "file:///path/to/history.file" -v 0
Versions File format information:
releaseDate | isLatest | releaseAgency | releaseId | releaseOrder | entityDescription
Sample record:
28-NOV-05 | false | http://nci.nih.gov/![]() | 05.10e | 26 | Editing of NCI Thesaurus 05.10e was completed on October 31, 2005. Version 05.10e was October's fifth build in our development cycle. |
| 05.10e | 26 | Editing of NCI Thesaurus 05.10e was completed on October 31, 2005. Version 05.10e was October's fifth build in our development cycle. |
<center>LoadOBO</center> |
Loads a file specified in the Open Biomedical Ontologies (OBO) format.
Options:
in,-input <uri> URI or path specifying location of the source file
-v, --validate <int> Perform validation of the candidate resource without loading data. If specified, the '-nf', -a' and '-t' options are ignored. Supported levels of validation include:
0 = Verify document is valid
nf,-noFail If specified, indicates that processing should not stop for recoverable errors
-a, --activate ActivateScheme on successful load; if unspecified the vocabulary is loaded but not activated
-t, --tag <id> An optional tag ID (e.g. 'PRODUCTION' or 'TEST') to assign.
Example: LoadOBO -in "file:///path/to/file.obo" -nf -a
LoadOBO -in "file:///path/to/file.obo" -v 0 |
<center>LoadOWL</center> |
Loads an OWL file. |
Note: Load of the NCI Thesaurus should be performed via the LoadNCIThesOWL counterpart, since it will allow more precise handling of NCI semantics.
Options:
in,-input <uri> URI or path specifying location of the source file
-v, --validate <int> Perform validation of the candidate resource without loading data. If specified, the '-nf', -a' and '-t' options are ignored. Supported levels of validation include:
0 = Verify document is well-formed
1 = Verify document is valid
nf,-noFail If specified, indicates that processing should not stop for recoverable errors
-a, --activate ActivateScheme on successful load; if unspecified the vocabulary is loaded but not activated
-t, --tag <id> An optional tag ID (e.g. 'PRODUCTION' or 'TEST') to assign.
Example: LoadOWL -in "file:///path/to/somefile.owl" -nf -a
LoadOWL -in "file:///path/to/somefile.owl" -v 0 |
<center>LoadRadLexProtegeFrames</center> |
|
<center>LoadUMLSSemnet</center> |
Loads the UMLS Semantic Network, provided as a collection of files in a single directory. The following files are expected to be provided from the National Library of Medicine (NLM) distribution:
|
Options:
in,-input <uri> URI or path of the directory containing the NLM files
-v, --validate <int> Perform validation of the candidate resource without loading data. If specified, the '-nf', -a' and '-t' options are ignored. Supported levels of validation include:
0 = Verify the existence of each required file
nf,-noFail If specified, indicates that processing should not stop for recoverable errors
-a, --activate ActivateScheme on successful load; if unspecified the vocabulary is loaded but not activated.
-t, --tag <id> An optional tag ID (e.g. 'PRODUCTION' or 'TEST') to assign.
-il --InheritanceLevel <int> If specified, indicates the extent of inherited relationships to import.
0 = none; 1 = all; 2 = all except is_a (default). All direct relationships are imported, regardless of option.
Example: LoadUMLSSemnet -in "file:///path/to/directory/" -nf --a --il 1
LoadUMLSSemnet -in "file:///path/to/directory/" -v 0 |
<center>LoadFMA</center> |
Imports from an FMA database to a LexEVS repository. Requires that the pprj file be configured with a database URN, username, password for an FMA MySQL based database. The FMA.pprj file and MySQL dump file are available at http://sig.biostr.washington.edu/projects/fm/ |
Options:
in,-input <uri> URI or path specifying location of the source file
-v, --validate <int> Perform validation of the candidate resource without loading data. If specified, the '-nf', -a' and '-t' options are ignored. Supported levels of validation include:
0 = Verify document is well-formed
1 = Verify document is valid
nf,-noFail If specified, indicates that processing should not stop for recoverable errors
-a, --activate ActivateScheme on successful load; if unspecified the vocabulary is loaded but not activated
-t, --tag <id> An optional tag ID (e.g. 'PRODUCTION' or 'TEST') to assign.
Example: LoadFMA -in "file:///path/to/FMA.pprj" -nf -a
or
LoadFMA -in "file:///path/to/FMA.pprj" -v 0 |
<center>LoadHL7RIM</center> |
Converts an HL7 RIM MS Access database to a LexGrid database |
in,-input <uri> URI or path specifying location of the source file
mf,-manifest <uri> URI or path specifying location of the manifest file
lp,-load preferences <uri> URI or path specifying location of the load preferences file
-v, --validate <int> Perform validation of the candidate
resource without loading data. If specified, the '-nf', -a' and '-t' options are ignored. Supported levels of validation include:
0 = Verify document is valid
nf,-noFail If specified, indicates that processing should not stop for recoverable errors
-a, --activate ActivateScheme on successful load; if unspecified the vocabulary is loaded but not activated
-t, --tag <id> An optional tag ID (e.g. 'PRODUCTION' or 'TEST') to assign.
Example: LoadHL7RIM -in "file:///path/to/file.mdb" -nf -a
or
LoadHL7RIM -in "file:///path/to/file.mdb" -v 0 |
<center>LoadMetaData</center> |
Loads optional XML-based metadata to be associated with an existing coding scheme. u,-urn <name> URN uniquely identifying the code system. Note: If the URN and version values are unspecified, a list of available coding schemes will be presented for user selection. |
Example: LoadMetadata -in "file:///path/to/file.xml" -nf -o
or
LoadMetadata -in "file:///path/to/file.xml" |
<center>LoadMrMap</center> |
Loads mappings file(s), provided in UMLS RRF format. Specifically MRMAP.RRF and MRSAT.RRF. |
Example: LoadMrMap -inMap "file:///path/to/MRMAP.RRF -inSat "file:///path/to/MRSAT.RRF" |
Special Batch Loading Functions
Shell Script |
Use and Function |
|---|---|
<center>LoadMetaBatch</center> |
Loads the NCI MetaThesaurus, provided as a collection of RRF files, using a batch loading strategy allowing a faster, more memory efficient load to occur. |
Options:
in,-input <uri> URI or path of the directory containing the NLM files
Example: LoadMetaBatch -in "file:///path/to/directory/" |
<center>LoadUMLBatch</center> |
Loads UMLS content, provided as a collection of RRF files in a
single directory. Files may comprise the entire UMLS distribution
or pruned via the MetamorphoSys tool. A complete list of
source vocabularies is available online athttp://www.nlm.nih.gov/research/umls/metaa1.html![]() .
.
Options:
in,-input <uri> URI or path of the directory containing the NLM files
s,-source vocabularies to load.
Example: LoadUMLSBatch -in "file:///path/to/directory/" -s "PSY" |
<center>ResumeMetaBatch</center> |
Resume a UMLS load. Loads will usually be restartable if they fail due to an
error. The loader will keep all loaded content and restart at the point of failure.
Options:
in,-input <uri> URI or path of the directory containing the NLM files
s,-source vocabulary to resume.
uri,-uri of vocabulary to resume.
version,-version of vocabulary to resume.
Example: ResumeMetaBatch -in "file:///path/to/directory/" -s "PSY" -uri "urn:123.4" -version "2.0" |
<center>ResumeUmlsBatch</center> |
Resume a UMLS load. Loads will usually be restartable if they fail due to an
error. The loader will keep all loaded content and restart at the point of failure.
Options:
in,-input <uri> URI or path of the directory containing the NLM files
s,-source vocabulary to resume.
uri,-uri of vocabulary to resume.
version,-version of vocabulary to resume.
Example: ResumeUmlsBatch.sh -in "file:///path/to/directory/" -s "PSY" -uri "urn:123.4" -version "2.0" |
Scheme and Metadata Removal
Shell Script |
Use and Function |
|---|---|
<center>RemoveScheme</center> |
Removes a coding scheme based on unique URN and version. |
Options:
u,-urn <urn> URN uniquely identifying the code system.
v,-version <versionId> Version identifier.
f,-force Force deactivation and removal without confirmation.
Example:
RemoveScheme -u "urn:oid:2.16.840.1.113883.3.26.1.1" -v "05.09e" |
<center>RemoveMetadata</center> |
Clears optionally loaded metadata associated with the specified coding scheme.
Options
u,-urn <urn> URN uniquely identifying the code system.
v,-version <id> Version identifier.
f,-force Force clear (no confirmation).
Note: If the URN and version values are unspecified, a
list of available coding schemes will be presented for
user selection.
Example: RemoveMetadata
Example: RemoveMetadata -u "urn:oid:2.16.840.1.113883.3.26.1.1" -v "05.09e" |
Pick List and Value Set Load Administration
Shell Script |
Use and Function |
|---|---|
<center>LoadPickListDefinition</center> |
Loads Pick List Definition content, provided in LexGrid canonical xml format. |
Options:
in,-input <uri> URI or path specifying location of the source file.
-v, --validate <int> Perform validation of the candidate
resource without loading data.
Supported levels of validation include:
0 = Verify document is well-formed
1 = Verify document is valid
Example: LoadPickListDefinition -in "file:///path/to/file.xml" |
<center>LoadValueSetDefinition</center> |
Loads Value Set Definition content, provided in LexGrid canonical xml format.
Options:
in,-input <uri> URI or path specifying location of the source file.
-v, --validate <int> Perform validation of the candidate
resource without loading data.
Supported levels of validation include:
0 = Verify document is well-formed
1 = Verify document is valid
Example: LoadValueSetDefinition -in "file:///path/to/file.xml" |
LexEVS Validation Test Script
Shell Script |
Use and Function |
|---|---|
<center>TestRunner*</center> |
Note: the LexEVS runtime and database environments must still be configured prior to invoking the test suite. Running any option other than -v will cause a large set of junits to be run
*
Located in
/test | Runs the test suite by invoking the Ant launcher.
Usage: TestRunner
-b, --brief
Run the LexBIG test suite and produce a text report with
overall statistics and details for failed tests only.
-f, --full
Run the LexBIG test suite and produce an itemized list of
all tests with indication of success/failure.
x,-xml
Run the LexBIG test suite and produce a report with extensive
information for each test case in xml format.
h,-html
Run the LexBIG test suite and produce a report suitable
for view in a standard web browser: this is the default
if no other option is specified.
v,-verify
Basic verification that LexEVS is configured properly
and basic systems are functioning.
Example: TestRunner -v
Example:
Run the full test suite of jUnits and save the report to an html file. (This can take quite some time.)
TestRunner --f -h
Run a verification of the LexEVS installation. A quick way to insure your system is up and running.
TestRunner -v |
Command Line Scripts and Wrappers Overview
LexEVS provides a set of shell scripts and code wrappers that provide command line users standard options and automatic printouts of current coding schemes when performing administrative functions using the command line.
Displaying Current Coding Schemes
When you have coding Schemes Loaded to LexEVS many command line script calls will bring up a printout of a table of the loaded coding schemes to be used as a selective menu for other functions.
Using optional parameters
A standard parameter interface allows users to customize command line calls to LexEVS. These options control things such as loader synchronicity, supplemental file locations, and fault conditions. Shell scripts contain comments on usage for each function wrappers options.
<br><nowiki>#</nowiki> Loads an OWL file. You can provide a manifest file to configure coding scheme <br><nowiki>#</nowiki> meta data. <br><nowiki>#</nowiki> <br><nowiki>#</nowiki> Options: <br><nowiki>#</nowiki> -in,--input <uri> URI or path specifying location of the source file <br><nowiki>#</nowiki> -mf,--manifest <uri> URI or path specifying location of the manifest file <br><nowiki>#</nowiki> -lp,--loaderPrefs<uri> URI or path specifying location of the loader preference file <br><nowiki>#</nowiki> -a, --activate ActivateScheme on successful load; if unspecified the <br><nowiki>#</nowiki> vocabulary is loaded but not activated. <br><nowiki>#</nowiki> -t, --tag <id> An optional tag ID (e.g. 'PRODUCTION' or 'TEST') to assign. <br><nowiki>#</nowiki> <br><nowiki>#</nowiki> Example: LoadOWL -in "file:///path/to/somefile.owl" -a <br><nowiki>#</nowiki> <br>java -Xmx3000m -XX:MaxPermSize=256M -cp "../runtime/lbPatch.jar:../runtime/lbRuntime.jar" org.LexGrid.LexBIG.admin.LoadOWL $@
Setting Java Virtual Machine Options
Heap size:
Memory management for heap size is a particular issue when loading larger Terminologies such as SNOMED or the NCI Thesaurus. Users need to be prepared to increase heap size if loads crash with heap size errors.
Example:
-Xmx:4000m
Permanent Generation:
LexEVS now depends on some Permanent Generation memory management during runtime. Typically set as follows:
-XX:MaxPermSize:256m
Installing sample vocabularies
This LexEVS installation provides a sample vocabulary, Automobiles.xml, that can be loaded into the database.
Step |
Action |
|---|---|
<center>1</center> |
In a Command Prompt window, enter <tt>*_cd |
/examples_*</tt> to go to the example programs. |
Figure 4 - Sample program output for finding properties and associations for a given code.
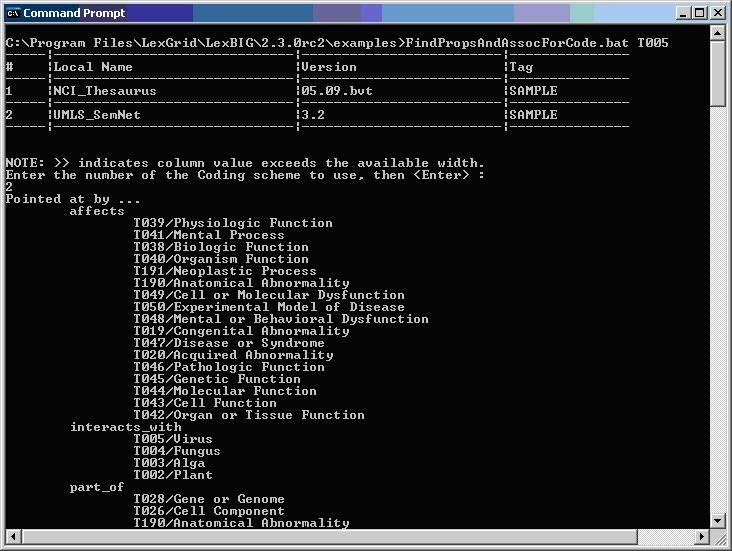
Figure 4 - Output of example programs using sample vocabulary
Installing NCI vocabularies
NCI Thesaurus vocabulary
This section describes the steps to download and install a full version of the NCI Thesaurus for the LexEVS Service.
border="0"
Step |
Action |
|---|---|
<center>1</center> |
Using a web or ftp client go to URL: ftp://ftp1.nci.nih.gov/pub/cacore/EVS/ |
 |
|
<center>2</center> |
Select the version of NCI Thesaurus OWL you wish to download. Save the file to a directory on your machine. |
<center>3</center> |
Extract the OWL file from the zip download and save in a directory on your machine. This directory will be referred to as NCI_THESAURUS_DIRECTORY |
<center>4</center> |
Using the LexEVS utilities load the NCI Thesaurus |
<tt>*_cd
/admin_*</tt>
For Windows installation use the following command
<tt>*_LoadOWL.bat --in "file:///
/Thesaurus_10.10d.owl"_*</tt>
For Linux installation use the following command
<tt>*_LoadOWL.sh --in "file:///
/Thesaurus_10.10d.owl"_*</tt> |
NOTE: |
<tt>The NCI Thesaurus has grown large enough that it can no longer be loaded on many typical desktop machines. We recommend a 64-bit operating system running on a multiprocessor computer with a minimum of 4g of memory. Server class Linux machines are the typical target for these loads. The time to load NCI Thesaurus will vary depending on machine, memory, and disk speed. Expect a couple of hours for a higher end machine</tt> |
|---|
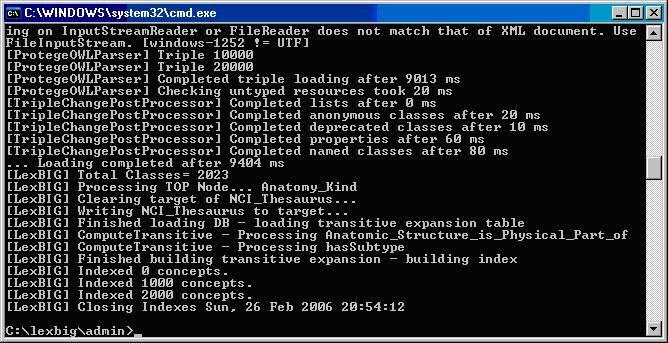
Table 7 – Example output from load of NCI Thesaurus 05.12f
...
LexBIG Processing TOP Node... Retired_Kind
LexBIG Clearing target of NCI_Thesaurus...
LexBIG Writing NCI_Thesaurus to target...
LexBIG Finished loading DB - loading transitive expansion table
LexBIG ComputeTransitive - Processing Anatomic_Structure_Has_Location
LexBIG ComputeTransitive - Processing Anatomic_Structure_is_Physical_Part_of
LexBIG ComputeTransitive - Processing Biological_Process_Has_Initiator_Process
LexBIG ComputeTransitive - Processing Biological_Process_Has_Result_Biological_Process
LexBIG ComputeTransitive - Processing Biological_Process_Is_Part_of_Process
LexBIG ComputeTransitive - Processing Conceptual_Part_Of
LexBIG ComputeTransitive - Processing Disease_Excludes_Finding
LexBIG ComputeTransitive - Processing Disease_Has_Associated_Disease
LexBIG ComputeTransitive - Processing Disease_Has_Finding
LexBIG ComputeTransitive - Processing Disease_May_Have_Associated_Disease
LexBIG ComputeTransitive - Processing Disease_May_Have_Finding
LexBIG ComputeTransitive - Processing Gene_Product_Has_Biochemical_Function
LexBIG ComputeTransitive - Processing Gene_Product_Has_Chemical_Classification
LexBIG ComputeTransitive - Processing Gene_Product_is_Physical_Part_of
LexBIG ComputeTransitive - Processing hasSubtype
LexBIG Finished building transitive expansion - building index
LexBIG Getting a results from sql (a page if using mysql)
LexBIG Indexed 0 concepts.
LexBIG Indexed 5000 concepts.
LexBIG Indexed 10000 concepts.
LexBIG Indexed 15000 concepts.
LexBIG Indexed 20000 concepts.
LexBIG Indexed 25000 concepts.
LexBIG Indexed 30000 concepts.
LexBIG Indexed 35000 concepts.
LexBIG Indexed 40000 concepts.
LexBIG Indexed 45000 concepts.
LexBIG Indexed 46000 concepts.
LexBIG Getting a results from sql (a page if using mysql)
LexBIG Closing Indexes Mon, 27 Feb 2006 01:44:22
LexBIG Finished indexing
NCI Metathesaurus vocabulary
Loading the Metathesaurus
This section describes the steps to download and install a full version of the NCI Metathesaurus for the LexEVS Service.
Step |
Action |
|---|---|
<center>1</center> |
Using a web or ftp client go to URL: ftp://ftp1.nci.nih.gov/pub/cacore/EVS/ |
 |
|
<center>2</center> |
Select the version of NCI Metathesaurus RRF you wish to download. Save the file to a directory on your machine. |
<center>3</center> |
Extract the RRF files from the zip download and save in a directory on your machine. This directory will be referred to as NCI_METATHESAURUS_DIRECTORY. Note: RELASE_INFO.RRF is required to be present for the load utility to work. |
<center>4</center> |
Using the LexEVS utilities load the NCI Thesaurus |
<tt>*_cd
/admin_*</tt>
For Windows installation use the following command
<tt>*_LoadMetaBatch.bat --in "file:///
/" _*</tt>
For Linux installation use the following command
<tt>*_LoadMetaBatch.sh --in "file:///
/"_*</tt> |
NOTE: |
<tt>NCI Metathesaurus contains many individual vocabularies some of which are large vocabularies in and of themselves. It requires many hours to load and index. It can require 36 hours on a multiprocessor machine with 6g plus memory. The total time to load NCI MetaThesaurus will vary depending on machine, memory, and disk speed. Because this loader uses a batch loading strategy it is less dependent on memory, but some users will see 3 or 4 day load times with average multiprocessor processing power.</tt> |
|---|
Resuming Loads
Since this loader is resource hungry we provide the option to restart should you find your resource settings to be inadequate. Resuming loads which have crashed or been interrupted by server problems is possible using the ResumeBatchLoad script set.
Step |
Action |
|---|---|
<center>1</center> |
Using the LexEVS utilities load the NCI Thesaurus |
<tt>*_cd
/admin_*</tt>
For Windows installation use the following command
<tt>*_ResumeMetaBatch.bat --in "file:///
/" -s "NCI Metathesaurus" -uri "urn:oid:2.16.840.1.113883.3.26.1.2" -version "200601"_*</tt>
For Linux installation use the following command
<tt>*_ResumeMetaBatch.sh --in "file:///
/" -s "NCI Metathesaurus" -uri "urn:oid:2.16.840.1.113883.3.26.1.2" -version "200601"_*</tt> |
NCI History
This section describes the steps to download and install a history file for NCI Thesaurus.
Step |
Action |
|---|---|
<center>1</center> |
Using a web or ftp client go to URL: ftp://ftp1.nci.nih.gov/pub/cacore/EVS/ |
<center>2</center> |
Select the version of NCI History you wish to download. Save the file to a directory on your machine. Select the VersionFile download to the same directory as the history file. |
<center>3</center> |
Extract the History files from the zip download and save in a directory on your machine. This directory will be referred to as NCI_HISTORY_DIRECTORY |
<center>4</center> |
Using the LexEVS utilities load the NCI Thesaurus |
<tt>*_cd
/admin_*</tt>
For Windows installation use the following command
<tt>*_LoadNCIHistory.bat --nf --in "file:///
" --vf "file:///NCI_HISTORY_DIRECTORY![]() }/VersionFile"_*</tt>
}/VersionFile"_*</tt>
For Linux installation use the following command
<tt>*_LoadNCIHistory.sh --nf --in "file:///
" --vf "file:///NCI_HISTORY_DIRECTORY![]() }/VersionFile" _*</tt> |
}/VersionFile" _*</tt> |
NOTE: |
<tt>If a 'releaseId' occurs twice in the file, the last occurrence will be stored. If LexEVS already knows about a releaseId (from a previous history load), the information is updated to match what is provided in the file. |
|---|
This file has to be provided to the load API on every load because you will need to maintain it in the future as each new release is made. We have created this file that should be valid as of today from the information that we found in the archive folder on your ftp server. You can find this file in the 'resources' directory of the LexEVS install.</tt> |
Deactivating and Removing a Vocabulary
This section describes the steps to deactivate a coding scheme and remove coding scheme from LexEVS Service.
Step |
Action |
||||
|---|---|---|---|---|---|
<center>1</center> |
Change directory to LexEVS administration directory Unknown macro: {LEXBIG_DIRECTORY}
/admin_*</tt> || <center>2</center> | Use the DeactiveScheme utility to prevent access to coding scheme. Once a coding scheme is deactivated, client programs will not be able to access the content for the specific coding scheme and version. | Tagging a Vocabulary
/admin_*</tt> |
||||
<center>2</center> |
Use the TagScheme to tag a coding system and version with a local tag name (e.g. PRODUCTION). This tag name can be used via LexEVS API for query restriction. |
Example:
<tt>TagScheme -u "urn:oid:2.16.840.1.113883.3.26.1.1" -v "10.10d" -t "PRODUCTION"</tt> |
IndexManagement
LexEVS indexes can be manually updated, removed and optimized by users. This is a useful function particularly when metadata, authoring and post processing may have taken place.
Remove Index
Step |
Action |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
<center>1</center> |
Change directory to LexEVS administration directory Unknown macro: {LEXBIG_DIRECTORY}
/admin_*</tt> || <center>2</center> | Use RemoveIndex script to remove an index for a particular coding scheme. |
/admin_*</tt> |
|||||||||||
<center>2</center> |
Use RebuildIndex script to rebuild an index for a particular coding scheme. |
Example:
Windows:
<tt>RebuildIndex.bat</tt>
Linux:
<tt>RebuildIndex.sh</tt> |
<center>3</center> |
Interactive commmand: <tt>Enter the number of the Coding Scheme to use, then <Enter>.</tt> User responds with column number for appropriate coding scheme. |
<center>4</center> |
Interactive command: <tt>REBUILD INDEX FOR URI? <uri displayed here> ('Y' to confirm, any other key to cancel).</tt> User responds with 'Y' |
<center>5</center> |
Command Line example: |
<center>6</center> |
Note: It may be useful and performance enhancing to run OptimizeLuceneIndex script after removing and rebuilding. |
Value Set and Pick List Loading
[Value Set and Pick List Scripts]