|
Page History
Applicable Releases:</strong> Up to caArray 2.X
<strong>Date entered:</strong> December 18, 2009
</span>
caArray Usage
questionDiv
question
Question: Working with Array Design Files in caArray questionEnd
| Scrollbar | ||
|---|---|---|
|
| Page info | ||||
|---|---|---|---|---|
|
...
| Panel | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| |||||||||
|
Topic: caArray Usage
Release: Up to caArray 2.X
Date entered: December 18, 2009
Question
Working with Array Design Files in caArray
Answer
Have a comment?
Please leave your comment in the caArray End User Forum.
| Scrollbar | ||
|---|---|---|
|
...
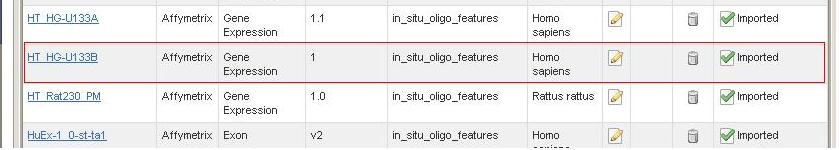
One of the most frequently asked questions from the new caArray users is: "Where can I obtain the array design file for my experiment?". Although the array providers' websites are still the best places to obtain the most updated array design files (For more details, see Where are the Array Design Files for caArray?), many array design files are now available to download at NCI's public caArray server, a new feature made available in caArray version 2.3.
...
| Panel |
|---|
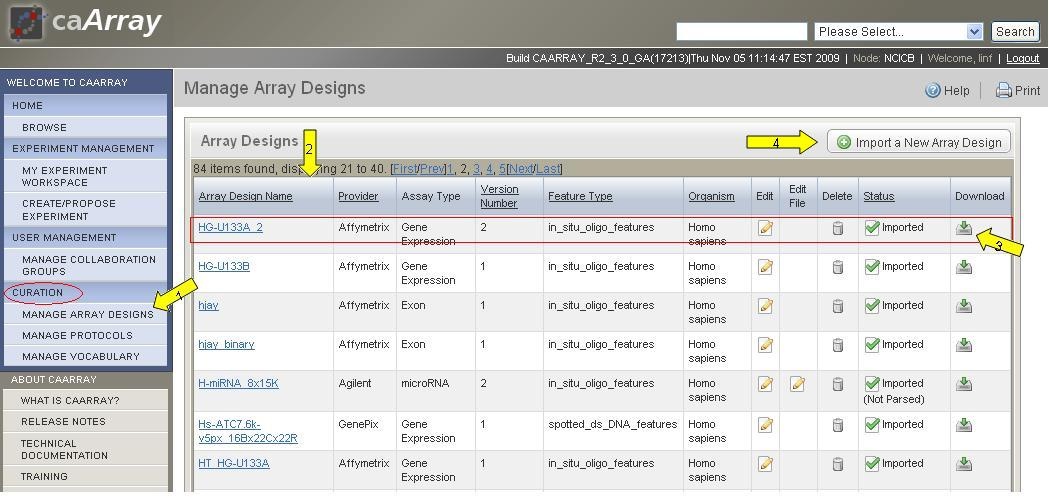
1. Click on "Manage Array Design" at the left menu section (Arrow 1). |
Upload a new array design file
...
| Panel |
|---|
*1. * Click on *Import a New Array Design *(Arrow 4 in Figure 1). |
| Panel |
|---|
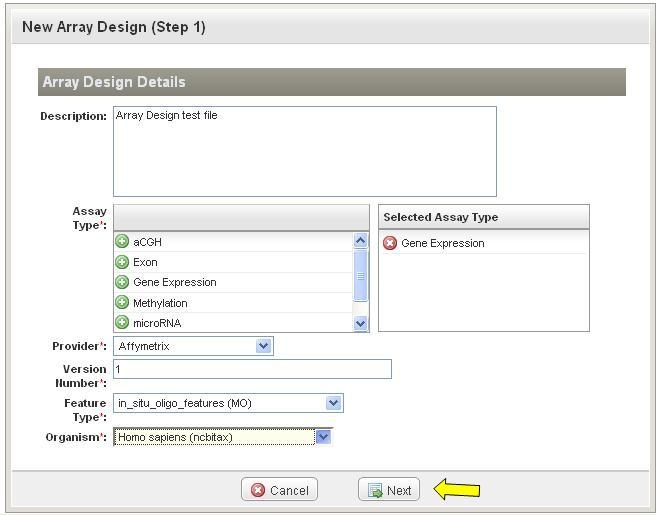
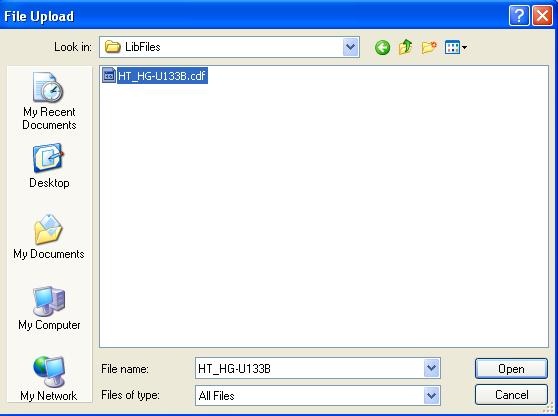
*4. * Click on "Next" to select an array design file interested.(Figure 3). |
| Panel |
|---|
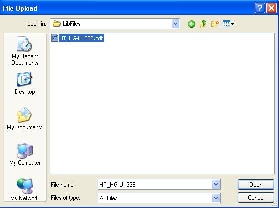
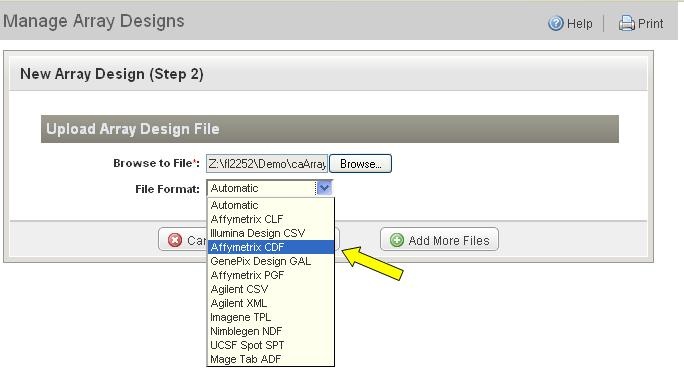
*5. * Identify the array design file format in the drop-down list (Figure 4). |
| Panel |
|---|
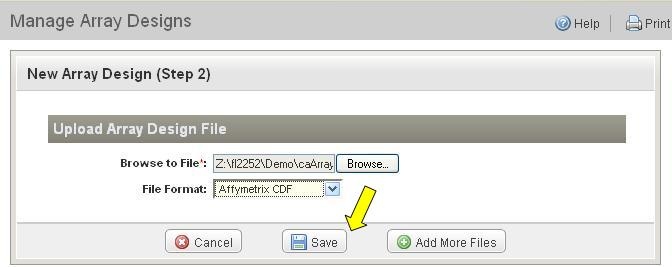
*6. * Multiple array design Files can be submitted to upload at the same time. Click on "Add More Files" to select a new file. After all of the files selected, submit the files to upload by clicking on "Save" (arrow in Figure 5). |
Validate an array design file
...
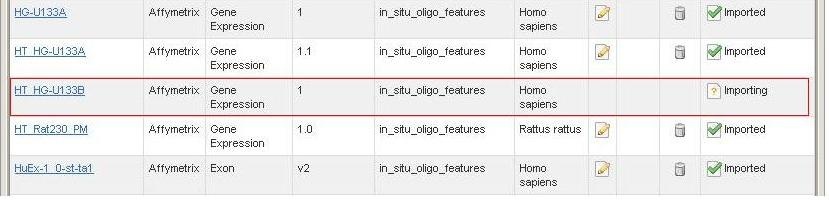
*2. * For more information on the different states of the array design files, refer to caArray Status of Importing: Imported vs Imported Not Parsed
Delete, Edit and Replace an array design file in caArray
...
After data has been imported into an experiment, the array design associated with that data cannot be removed from the experiment's list of array designs. Conversely, once an array design has been imported, the file associated with it cannot be changed. Array designs that are not associated with an experiment can now be deleted. To delete an array design, follow this step:
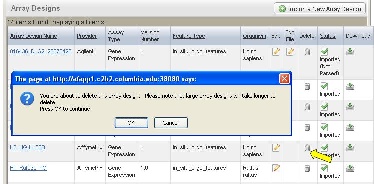
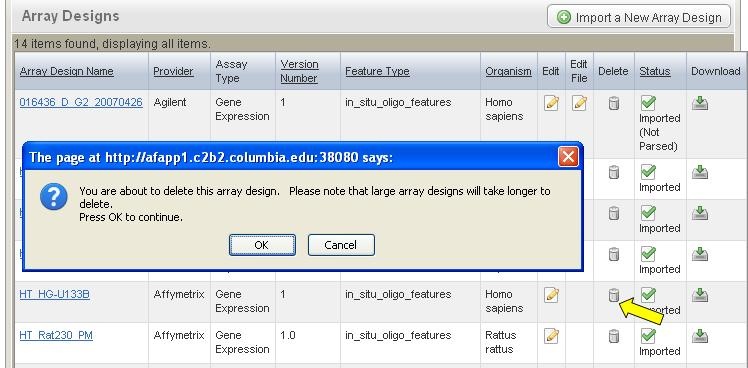
*1. * On the row corresponding to the array design, click the Delete icon (Arrow in Figure 7).
*2. * A confirmation window will pop up. Once you click the "OK" button, the file will be physically deleted from caArray.
Edit an Array Design
...
1. On the row corresponding to the array design, click the Edit icon (Figure 8).
2. The required fields become editable; enter any edits.
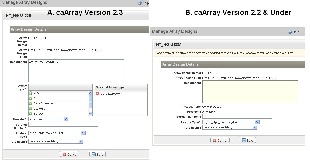
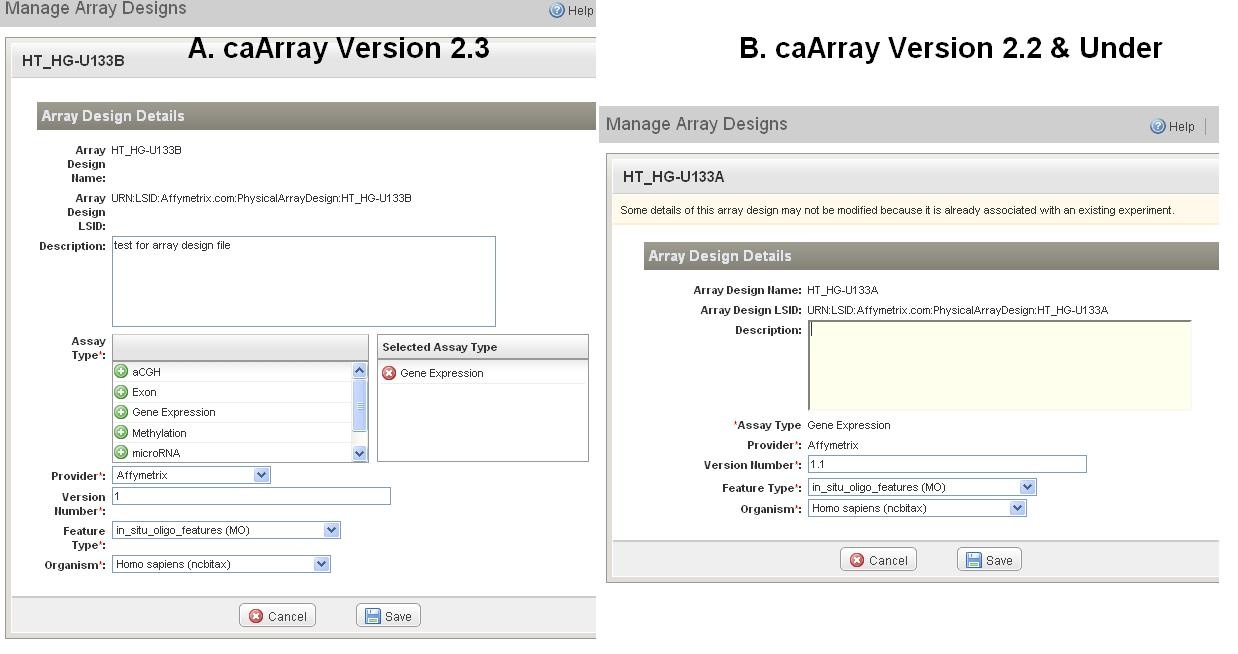
3. Please note, the editable fields are limited for the array design files uploaded prior to caArray 2.3. Starting from caArray 2.3, all required fields become editable (Figure 9).
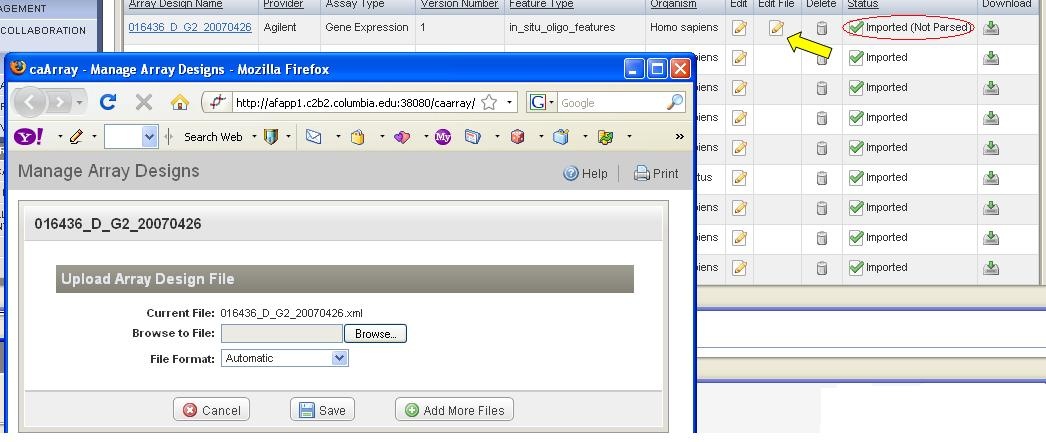
Replace an Array Design
...
| Panel |
|---|
*1. * Find the file that is replaceable (the file with a status of "Imported (Not Parsed)" (circle in red in Figure 10). |
Have a comment? Please leave your comment in [ caArray End User Forum|http://cabig-kc.nci.nih.gov/Molecular/forums/viewtopic.php?f=6&t=436]
...