|
Page History
| Table of Contents |
|---|
Released by / Effective Date:
Written by: | ||
Printed Name: | Title: | Signature/Date: |
Michal Lijowski | ||
Approved by: | ||
Printed Name: | Title: | Signature/Date: |
QA Approved by: | ||
Printed Name: | Title: | Signature/Date: |
PURPOSE
Curation of biomedical information is accomplished by selecting relevant publications, extracting reported text and data, submitting extracted information into ISA-TAB-Nano and caNanoLab, and keeping track of performed activities.
SCOPE
REFERENCES
RESPONSIBILITIES
DEFINITIONS
REAGENTS, MATERIALS AND EQUIPMENT
HEALTH AND SAFETY CONSIDERATIONS
PROCEDURE PRINCIPLES
Summary
Curation of biomedical information is accomplished by selecting relevant publications, extracting reported text and data, submitting extracted information into ISA-TAB-Nano and caNanoLab, and keeping track of performed activities.
...
- The NCI collaborator provides a list of publications suggested for curation.
- Publications are evaluated whether they are curable, i.e. comprise information relevant for curation in caNanoLab.
- Data extraction from the publication.
- Submission of extracted data into caNanoLab and into ISA-TAB-Nano forms.
- Information submitted into caNanoLab must be machine-readable, searchable and comply with the established standards.
- Sending to authors of a publication, a request for additional data and submission of provided additional data into caNanoLab and ISA-TAB-Nano forms.
Initial Steps
- Create a caNanoLabData folder on a system or server that gets backup regularly. The caNanoLabData folder contains folders, named after institution or collaboration, e.g. USC_UV which contain additional subfolders. These subfolders, e.g., are named after first author of a publication, publication abbreviation, and publication year, e.g. , JCrecente-CampoJCR2019 contain an individual publication, i.e. PDF file and any supplemental data associated with a publication, extracted data, and supplemental data provided by an author. The caNanoLabData folder contains all auxiliary files, e.g. a list of cell lines, a list of all curated publications, a list of chemical compounds, a list of new terms, and recently added list of Bioportal terms.
- Create a subfolder in caNanoLab folder to store the publication and extracted data. A subfolder name comprises first author name, journal name, and year of publication. Additional subfolder within this subfolder is created to store ISA-TAB-Nano forms.
Extract Data
- Establish a number of samples, which have different compositions or properties, and a number of characterizations using the information provided in the text, tables, figures, and figures' captions in curated publication.
- Establish sample names, following the pattern: abbreviation(s) of institution names, name of the first author (without a middle name), a custom abbreviation of the journal title, year of publication, and sample sequential number, e.g. USC_UV-JCrecente-CampoJCR2019-01.
- Associate samples with characterizations based on information provided in the text, tables, figures, and figures’ captions. This information is kept in a text file listing all samples and associated characterizations (Figure 1).
- Extract information on the composition, physicochemical, in vitro, and in vivo characterizations, numerical data for each individual sample into a corresponding text file. Replace Greek fonts with English equivalents e.g. α replace with alpha. In units replace μ with u. Check the extracted text for nonstandard hyphens. Remove references to figures and to publications. Rephrase active sentences to passive. For example “We synthesized the previously reported μMOF, Hf-DBA (DBA = 2,5-di(p-benzoato)aniline), and used it as a control.” replace with “The previously synthesized and reported μMOF, Hf-DBA (DBA = 2,5-di(p-benzoato)aniline), was used as a control”.
Establish definitions for new terms used in the publication, which are not in the caNanoLab glossary or Bioportal, but in other sources, like Wikipedia, and references therein, Encyclopedic Dictionary of Genetics, Genomics, and Proteomics. Record this definition or term, in a designated text file or if possible enter into caNanoLab, e.g. in targeting functionalized entity, a new target, i.e. gene.
If the information provided by the publication on e.g. name of an instrument or a chemical compound does not agree with the information provided somewhere else, like manufacturer catalog, retain for curation information provided by the publication and record a discrepancy for correspondence with authors, in a file, which contains a request for numerical data which were used to generate figures.
Info icon false Crecente-Campo J, Guerra-Varela J, Peleteiro M, Gutierrez-Lovera C, Fernandez-Marino I, Dieguez-Docampo A, Gonzalez-Fernandez A, Sanchez L, Alonso MJ. The size and composition of polymeric nanocapsules dictate their interaction with macrophages and biodistribution in zebrafish. J Control Release. 308:98-108 (2019).
1 biopolymer (inulin) small nanocapsule
physicochemical size zeta potential Figure 1
in vitro cytotoxicity Figure 2
in vivo stability Figure S1 toxicity Figure 4 survival Table S1 Table S2 Table S3
2 biopolymer (inulin) medium nanocapsule
physicochemical size zeta potential Figure 1
in vitro cytotoxicity Figure 2
in vivo stability Figure S1 toxicity Figure 4 survival Table S1 Table S2 Table S3
3 biopolymer (chitosan) small nanocapsule
physicochemical size zeta potential Figure 1
in vitro cytotoxicity Figure 2
in vivo stability Figure S1 toxicity Figure 4 survival Table S1 Table S2 Table S3
4 biopolymer (chitosan) medium nanocapsule
physicochemical size zeta potential Figure 1
in vitro cytotoxicity Figure 2
in vivo stability Figure S1 toxicity Figure 4 survival Table S1 Table S2 Table S3
5 biopolymer (inulin) fluorescent small nanocapsule
in vitro targeting cell internalization Figure 3
in vivo biodistribution Figure 5 biodistribution Figure 6 biodistribution Figure S3 biodistribution Figure S4 biodistribution Figure S5
6 biopolymer (inulin) fluorescent small nanocapsule
in vitro targeting cell internalization Figure 3
in vivo biodistribution Figure 5 biodistribution Figure 6 biodistribution Figure S3 biodistribution Figure S4 biodistribution Figure S5
7 biopolymer (chitosan) fluorescent small nanocapsule
in vitro targeting cell internalization Figure 3
in vivo biodistribution Figure 5 biodistribution Figure 6 biodistribution Figure S3 biodistribution Figure S4 biodistribution Figure S5
8 biopolymer (chitosan) fluorescent medium nanocapsule
in vitro targeting cell internalization Figure 3
in vivo biodistribution Figure 5 biodistribution Figure 6 biodistribution Figure S3 biodistribution Figure S4 biodistribution Figure S5
Figure 1. A typical text showing associations between samples and characterizations.
Submit caNanoLab Data
Submit extracted information and reported numerical data into caNanoLab following online caNanoLab User Guide, which is accessible by selecting caNanoLab FAQ or Online Help buttons (Figure 2). If submitting a new term in any field in caNanoLab use lowercase.
- Login into caNanoLab and after successful login select either SAMPLES tab in the top bar or Submit Samples button (Figure 2).
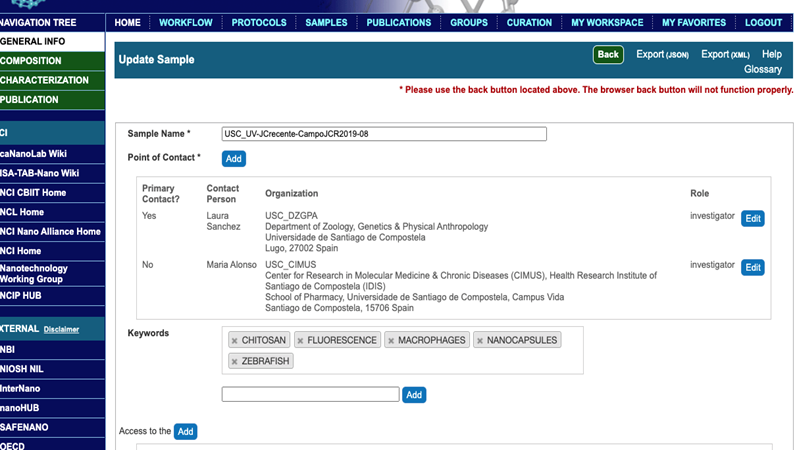
Figure 2. caNanoLab home after login. Selecting tabs allows search samples, protocols, publications, and submission of samples, protocols, and publications. - Submit sample name, contact information, i.e. custom generated abbreviation for institution name(s), role (either manufacturer or investigator), and addresses of corresponding authors, first name, middle initial, last name, phone number, email address, and keywords relevant to the publication, into General Info section. The first author is a primary point of contact (Figure 3).
Figure 3. A General Info window after submission of relevant data. Selecting Composition, Characterization, or Publication buttons on top left allows submission of sample composition, its characterizations, and the corresponding publication citation. - Composition submission.
- Select Composition button below General Info (Figure 3).
- Select nanoparticle entity type from Nanoparticle Entity Type drop down menu.
- Particle description into Description field.
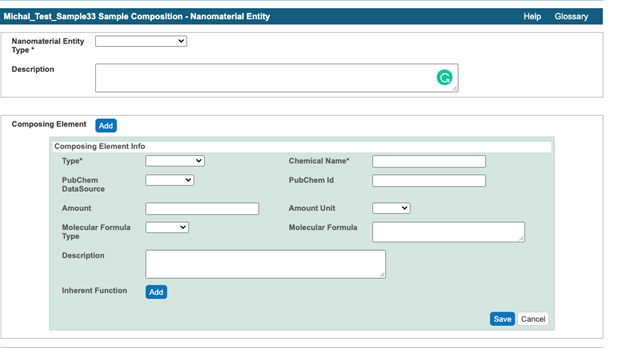
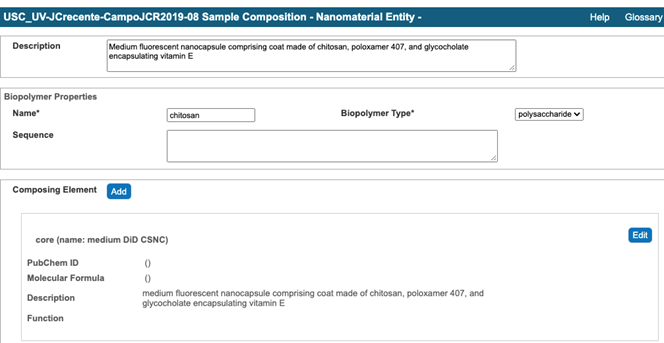
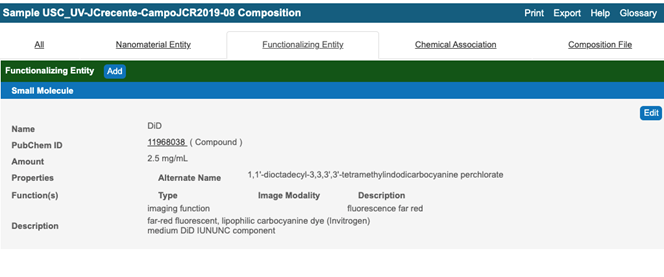
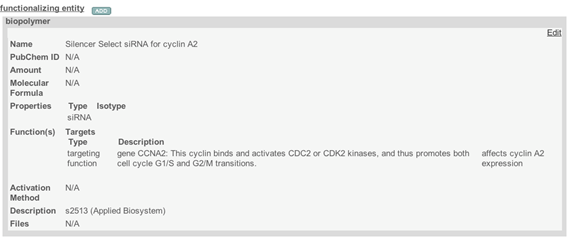
- Submit sample composition into Nanomaterial entity section. This includes chemical name of sample component, its type from a drop down menu, its full name in the description field, PubChem ID, , and amount. If any of sample components has a function, e.g. targeting, the information is indicated in Inherent Function field (Figure 4). At the time of writing of this document (caNanoLab 2.3.10) the inherent function does not work properly. It is advised to use Functionalized Entity section to submit Inherent Function information (Figure 6, Figure 7). In addition, there only three standard functions in the drop down menu, namely imaging function, targeting function, and therapeutic function. The very first Composing Element comprises information about a whole sample (Figure 5).
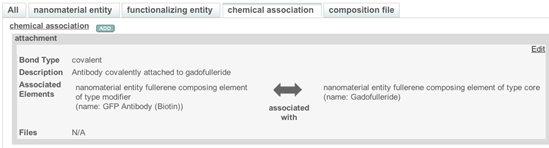
- If information about a link between functionalized entity and samples components is reported in the publication, then this information is entered into chemical association section (Figure 6).
Figure 4. A window for submission of information about sample constituent
Figure 5. An example of the first composing element
Figure 6. A typical window of functionalized entity
Figure 7. A typical window for a targeting functionalized entity
Figure 8. A typical chemical association window
- Characterization submission.
- Select Characterization button (Figure 3).
- Select appropriate Characterization Type, i.e. select, either physicochemical, in vitro, or in vivo from a drop down menu (Figure 9).
- Select from a drop down menu a Characterization Name. If a corresponding Characterization Name or Assay Type are not available, select either other_pc as physicochemical Characterization Name, other_vt for in vitro Characterization Name, other_vv for in vivo Characterization Name, or other_ex_vv for ex vivo Characterization Name.
- Select an assay type or enter a new assay type in the Assay Type field drop down menu, if an appropriate assay type is not available.
- Select Characterization Source from a drop-down menu.
- Enter a cell line, if a field for a cell line exists.
- Enter an assay description into a Design and Methods Description field.
- Enter technique(s) and instrument(s), used in an assay, either by selecting existing technique and instrument or by adding a new technique and instrument into respective drop down menus.
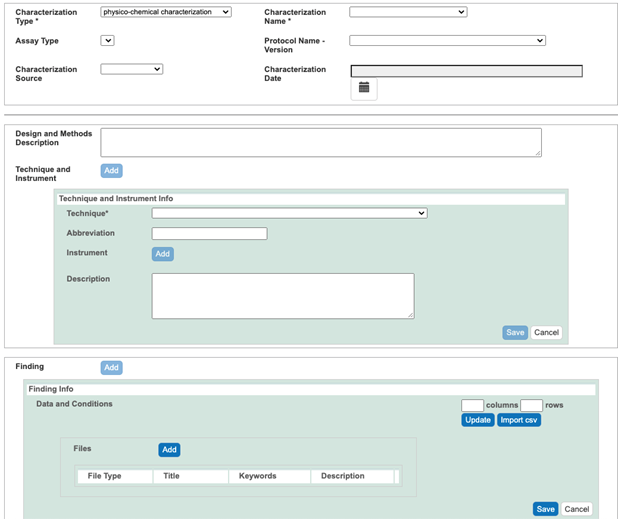
- If numerical data are available, click the Findings button, then in case a small amount of data, enter number of columns and rows required to accommodate these data, select Update button, annotate columns, and enter data. In case of large amount of data create a csv file, select Import cvs button, and select a csv file to import. Regarding columns annotation, first one have to select Column Type, either condition or datum. If e.g. numerical data are provided as mean, uncertainty, and number of replications than first Column Name (mean) can be retrieved from drop menu if the name associated with this column exists in the menu, otherwise one have to select other on the bottom of the menu and enter a new corresponding Name into New Column Name field, select Column Value Type (mean), select Column Value Unit. One have to do the same for column holding uncertainties, and number of replications. In case Column Name exists in the drop down menu, one can use the name in in the menu for this first column (mean), but for second and third column one have to proceed above, by selecting other on the bottom of the menu, entering existing Column Name, Column Value Type, and Column Value Unit.
- Submit the description of the results from an assay into an Analysis and Conclusion field (Figure 10).
Figure 9. A Physicochemical Characterization window
Figure 10. A Physicochemical Characterization window with submitted data
- Publication submission.
- Select Publication tab (Figure 3).
- Select a Publication Type for a drop-down menu.
- Select Publication Status from a drop-down menu.
- Enter PubMed ID and click outside PubMed ID field to obtain a citation for this publication (Figure 8).
- If the publication does not exist in PubMed, then enter the publication DOI, its title, journal name, year of publication, volume, start and end pages, list of author names, keywords, abstract in Description field.
- Select Research Categories.
- Associate the publication citation with submitted samples as follows.
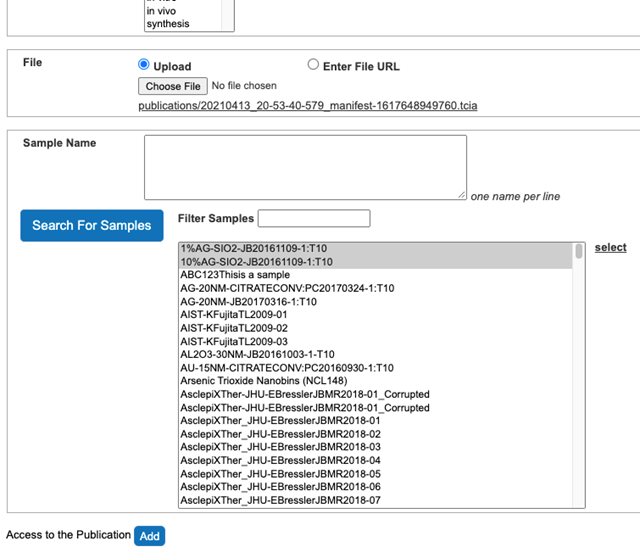
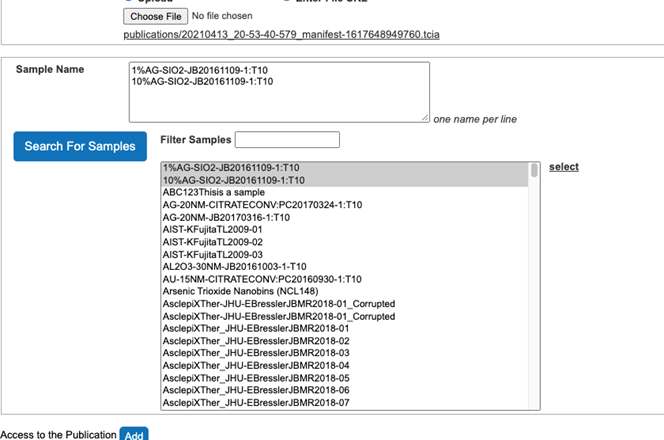
- Select Search For Samples button.
- Select Samples associated with the publication from the list of all samples (Figure 11).
- Click select button on the right side to associate Samples with the publication (Figure 12).
Figure 11. Samples association with the publication
Figure 12. Samples are associated with the publication. - Set access to the publication citation as “public”.
- Review entries submitted into caNanoLab for consistency with information in the curated publication. Correct any issues.
- Make all samples “public”.
Submit ISA-TAB-Nano Data
At the time of writing of this document, the curation in ISA-TAB-Nano follows the ISA-TAB-Nano 1.3 Release. More detailed information is provided in ISA-TAB-Nano wiki https://wiki.nci.nih.gov/display/icr/isa-tab-nano. The ISA-TAB-Nano forms filenames comprise a prefix corresponding to a specific form, i.e. i_ for an investigation form, s_ for a study form, a_ for an assay form, m_ for a material form, a custom abbreviation of institution(s) names, a name of the first author (first name abbreviation, full last name), a custom abbreviation of journal title and a year of publication, e.g. i_USC_UV-JCrecente-CampoJCR2019. A suffix for Study indicates a study type e.g. physicochemical, in_vitro. A suffix for Assay file in addition to a type of Study is related to, includes a name of assay, e.g. size, zeta potential.
...
a_ USC_UV-JCrecente-CampoJCR2019-physicochemical-size-DLS.
Investigation Form
The very first lines of Investigation form (Figure 13) are dedicated to names of ontologies from Bioportal (http://bioportal.bioontology.org). The information about ontologies is added, while creating ISA-TAB-Nano Investigation form and selecting appropriate annotation from Bioportal for terms, which are entered into ISA-TAB-Nano forms. In case, a term exists in multiple ontologies, then the most in depth annotation is selected.
...
Figure 14. A general information section of Investigation form.
Study Section of Investigation Form
Based on information obtained earlier and related to sample characterizations, identify studies and assays, which are common to a specific study.
...
Figure 19. A Study Contacts section
Material Form
Create a number of Material forms corresponding to a number of identified samples. In addition to identified samples, which are submitted into caNanoLab, one can create Material forms for substances, like sucrose, saline, or drugs, which are used as control materials.
...
Figure 20. Part of Material form showing a linkage subsection
Study and Assay Forms
The number of created Study forms should be equal to the number of Study sections in the Investigation form. In the case of imaging studies, create corresponding Study and Assay forms, when images are available without any restriction. Then, the name of file containing the image is entered into an Image File field of a corresponding Assay form.
Study Form
The number of fields in a Study form and which fields are in this form depends on the number of Study Factors, which are entered in the Investigation form for this particular Study. The common fields in Study form are in all types of Studies are Source Name, Sample Name, Factor and Parameter Values (Figure 18), if both latter are specified in the Investigation form.
- Enter into Source Name, either Material Source Identifier in case of physicochemical Study e.g. USC_UV-JCrecente-CampoJCR2019-04, cell line in case of in vitro characterization e.g. H-1650, or animal name e.g mice in case of in vivo
- Enter material type, either as a free-text description or as a term from Bioportal, its Term Accession Number, and Term Source REF.
- Enter into Characteristics field cell type and cell line in case of in vitro Study or animal type in case of in vivo
- Enter Factor Value, e.g. Material Source Identifier in case, in which an experiment involved only different samples, or entered multiple Factor values. In Sample Name field, enter either name of nanomaterial sample in case, there no Factors, or a name, which includes a combination of sample name, factors numerical values, cell name, and/or animal name. Number of lines in Study form depends on number of Factor Values and numerical Factor Values.
- Enter protocol name from the corresponding Study Section in the Investigation form into Protocol REF field.
- Enter Study filename into the corresponding Study File Name field in the Investigation form.
Figure 21A. Part of a Study form showing primary entries in this form for in vitro assay
Figure 21B. Part of a Study form showing entries in this form for in physicochemical assay
Assay Form
For each study, create a number of Assays forms corresponding to assays in the Study Assay subsection of the Study section in the Investigation form, in case corresponding numerical data are readily available. Enter the following into an Assay form (Figure 19).
...
Figure 22. Part of Assay form showing important entries
Final Steps
- Complete the Investigation form by entering Material File Names, Material Source Name, Study File Names, and Assay File Names.
Convert all files into csv format. This step can be performed in macOS and Linux platforms using the unoconv script.
unoconv -e FilterOptions=9/32,,9 -f csv -o ../ISA-TAB-Nano_csv *.xlsx if ISA-TAB-Nano forms were created using the Excel.
...
- In the Investigation form replace all extensions e.g. xlsx with csv using a text editor or a script.
- Compress all forms and associated information into a single compressed file and post the file into the caNanoLab Data Curation Project Wiki under ISA-TAB-Nano Curated Examples including a citation of the publication.
- Update caNanoLab/ISA-TAB-Nano curation status in the caNanoLab Data Curation Project Status file.
- After completing caNanoLab curation provide NCI collaborator with explicit list and description of data needed from investigators to complete curation task.
- Submit to the NCI JIRA tracker reports regarding defects encountered in caNanoLab and requests for improvements.
- Prepare and disseminate weekly report to the Government Sponsor, Project Officer, Leidos Technical Project Manager (COB Friday), and ESI Manager.
- Attend weekly teleconference.
- Prepare and disseminate the monthly report.
ATTACHMENTS
REVISION HISTORY
Version | Change | Reason |
1.0 | Currently no procedure; new initiative requiring communication of expectations. |