|
Page History
...
| Info | ||
|---|---|---|
| ||
Setting survival values is optional if you do not plan to use the K-M plot analysis feature or if you do not have this kind of data (survival values) in the file. |
For some applications, such as REMBRANDT and I-SPYIn caIntegrator, survival values are not pre-defined in the databases when you load the data. In caIntegratorHowever, however, you can review and define survival value ranges in a data set you are uploading to a study. To be able to do so, you need to understand the kind of data that can comprise the survival values.
...
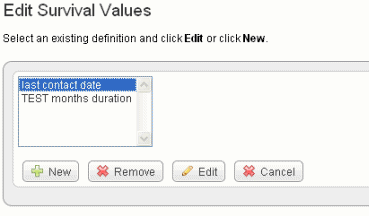
- On the Edit Study page, click Edit Survival Values. This opens the Survival Value Definitions dialog box, shown in the following figure.
- Click New to enter new survival value definitions.
OR
Click Edit to edit existing survival value definitions.
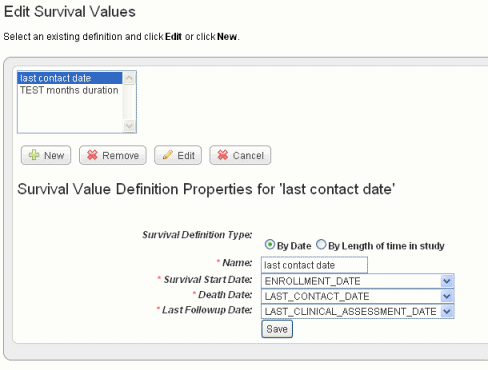
The dialog box extends, now displaying radio buttons and three drop-down lists that show column headers for date metadata in the spreadsheet you have uploaded. If survival value ranges have already been added to a study, they display on the page. The dialog box is shown in the following figure.
- Survival values can be defined by Date or by Length of time in the study. Select the radio button for the category that defines your survival the data.
- In the drop-down lists, select the appropriate survival value definitions for each field listed. You might want to refer to the column headers in the data file itself. Dates Because dates covered by the definitions are already in the data set. You , you cannot enter specific dates.
Field Type
Description
Survival Definition Type
Select whether the survival time is defined by dates or length of time subject was in the study.
Name
Enter a unique name that adequately describes the survival values you are defining here. Example: Survival from Enrollment Date or Survival from Treatment Start. The name you enter displays later when you are selecting survivals to create the K-M plot.
Survival Length Units
Select the appropriate units for this data.
Survival Start Date
Select the column header for this data.
Death Date
Select the column header for this data.
Last Followup Date
Select the column header for this data.
...
Once you have loaded subject annotation data and identified subject IDs, you can add one or more sets of array genomic sample data from caArray , which to the study. caIntegrator maps the data by sample IDs to the patient subject IDs in the subject annotation data, covered in this section, or you can load imaging files from NBIA, also mapped by IDs to the patient subject data. This is discussed in #Working with Imaging Data. You can also edit genomic data information that you have already added to the study. Genomic sample data and imaging data are independent of each other, so neither is required before loading the other.
...
- On the Edit Study page where you have selected and added the subject annotation data, click the Add New button under Genomic Data Sources. You can upload genomic data only from caArray.
This opens the Edit Genomic Data Source dialog box. Enter the appropriate information in the fields, shown in the following figure . The fields are and described below.
Field Types
Field Description
caArray Web URL
Enter the URL for the caArray instance to be used for the genomic data sources. This will enable a user to link to the referenced caArray experiment from the study summary page.
caArray Host Name
Enter the hostname for your local installation or for the CBIIT installation of caArray, . If you misspell it, you will receive an error message.
caArray JNDI Port
Enter the appropriate server port. See your administrator for more information. Example: For the CBIIT installation of caArray, enter 8080.
caArray Username and caArray Password
If the data is private, you must enter your caArray account user name and password; you must have permissions in caArray for the experiment. If the data is public, you can leave these fields blank.
caArray Experiment ID
Enter the caArray Experiment ID which you know corresponds with the subject annotation data you uploaded. Example: Public experiment "beer-00196" on the CBIIT installation of caArray (array.nci.nih.gov). If you misspell your entry, you will receive an error message.
Vendor
Select either Agilent or Affymetrix.
Data Type
Select Expression or Copy Number.
Platform
If appropriate, select the Agilent or Affymetrix platform.
Info title Note Because you can add more than one set of genomic data to a study, a study can also have multiple platforms, one for each set of genomic data.
Central Tendency for Technical Replicates
If more than one hybridization is found for the reporter, the hybridizations will be represented by this method.
Indicate if technical replicates have high statistical variability
If more than one hybridization is found, checking this box will display a ** in the genomic search results when a reporter value has high statistical variability.
Standard Deviation Type
When the checkbox for indicating if technical replicates have high statistical variability is checked, this parameter becomes available. Select in the drop-down the calculation to be used to determine whether or not to display a ** (see previous bullet point).
--Relative- calculates the Relative Standard Deviation in percentage value
--Normal- calculates the Standard Deviation in numeric valueStandard Deviation Threshold
When the checkbox for indicating if technical replicates have high statistical variability is checked, this parameter becomes available. This is the threshold at which the Standard Deviation Type is exceeded and the reporter is marked with a **.
- Click Save.
...
- Start with the 6-column mapping file template, described as follows:
- All platforms – Raw (level 1) data cannot be mapped; only normalized, processed (level 2) data is acceptable.
- The required six-column file format uses the following columns:
- Subject ID
- Sample ID
- Name of supplemental file (if appropriate, as attached to the experiment in caArray)
- Probe Header – Name of column header (in the supplemental file) which contains the probe IDs.
- Value Header – Name of column header (in the supplemental file) which holds the level 2 data.
- Sample Header – Name of column header (in the supplemental file) which holds the level 2 data.
Info title Note Only one of the last 2 columns is used: a single sample per file uses the Value Header column; multiple samples per file used Sample Header column. Unused columns are blank.
The following figure shows an example multiple sample mapping file in CSV format.
- When you use the mapping file, make sure you use the subject ID for mapping. If the file is human data, the subject ID is the patient ID.
- Determine whether your data in caArray is "imported and parsed" or "supplemental". These are the 'Loading Types' referred to in Step 4 of #Steps for Mapping Genomic Data. Fill in the 6-column mapping file according to the following standard:
- Imported and parsed – Complete only the first two columns of the 6-column mapping file as described above. You can ignore the remaining columns.
- Supplemental – Supplemental data comes in two flavorstypes: "single sample per file" and "multiple samples per file". Only In either case, only one of the last two columns is used. If the supplemental data format is: Single , single sample per file – , the column named "Sample_Header" can be left empty. Multiple If the supplemental data format is multiple samples per file – , the column named "Value_Header" can be left empty.
The following steps use data of either type.Info title Note Supplemental files from caArray for mapping data must be configured appropriately. For information, see Supplemental Files Configuration.
Steps for Mapping Genomic Data
...
- On the Edit Study page, click the Map Samples button. This opens the Edit Sample Mappings page, shown in the following figure.
- The first two caArray fields may be populated with the information for the instance of caArray to which you have access. You can, however, enter the caArray information described in the following table, if appropriateyou prefer.
Field
Description
caArray Host Name
Enter the hostname for your local installation or for the CBIIT installation of caArray, . If you misspell it, you will receive an error message.
caArray JNDI Port
Enter the appropriate server port. See your administrator for more information. Example: For the CBIIT installation of caArray, enter 8080.
caArray Username
Enter your caArray account user name and password; you must have permissions in caArray for the experiment if it is private. If the data is public, you can leave this field blank.
caArray Experiment ID
Enter the caArray Experiment ID which you know corresponds with the subject annotation data you uploaded. Example: Public experiment "beer-00196" on the CBIIT installation of caArray (array.nci.nih.gov). If you misspell your entry, you will receive an error message.
- Enter the Loading Type of the data file you plan to map. (File types are described in #Creating a Mapping File).
- In the Subject to Sample Mapping File section, click Browse to navigate for the Sample Mapping CSV file that you created (described in #Creating a Mapping File). This provides caIntegrator with the information for mapping patients to caArray samples.
- Click the Map Samples button.
If the caArray data you have identified is imported and parsed, when you click the Map Samples button, the mapping takes place as the data is uploaded into caIntegrator. If the caArray data is supplemental, the mapping does not occur until the study is deployed. - Mapped samples will be listed in the Samples Mapped to Subjects section; scroll down the page to view them (see the following figure). Unmapped samples show at the top of the caIntegrator page. They were loaded from caArray, but they are not in the mapping file. These are not used for integration.
Info title Note If you have already mapped samples, when you first open this page they are listed in the Samples Mapped to Subjects section. If you have not already mapped samples, all of the samples in the caArray experiment you selected are listed as unmapped, because caIntegrator does not know how these sample names correlate to the patient data in the subject annotation file until you upload the subject to sample mapping file.
Uploading Control Samples
A Control Samples file is used to calculate fold change data, which compares "tumor" sample gene expression in the caArray experiment to the control samples to identify those that exhibit up or down gene regulation. Control samples can be the "normal" samples, but that is not necessarily always the case.
To upload the control samples, follow these steps:
...
| Info | ||
|---|---|---|
| ||
If a Control Set is to be used in Gene Expression For Annotation, or Gene Expression plots Plots for Annotation Query, then the control set should be composed of only samples which are mapped to subjects. |
Configuring Copy Number Data
You can add copy number data for a genomic data source by uploading the mapping file. This allows you to configure parameters to be used when segmentation data is being configured.
...
- In the Genomic Data Sources section of the Edit Study page, for the data you have already added, click Configure Copy Number Data button.
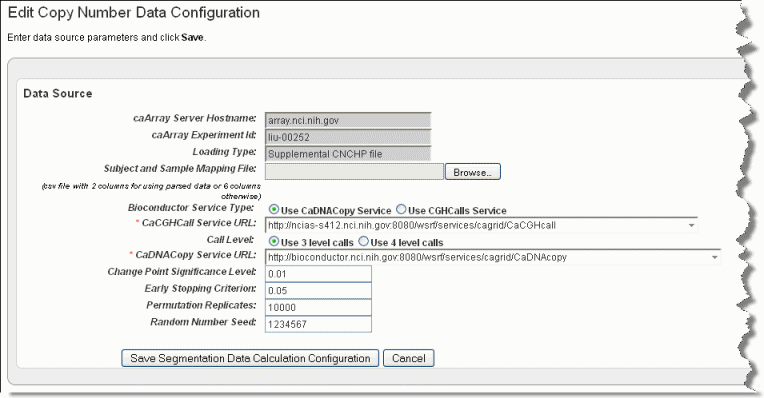
The Edit Copy Number page, shown in the following figure, opens.Info title Note This link is available only if you have uploaded copy number data and you are configuring a Copy Number data type (as indicated by the Data Type column on the Edit Study page).
- Browse for and enter appropriate information to identify and retrieve the copy number mapping file. The fields are described in the following table. An asterisk indicates a required field.
Field
Description
caArray Service Host Name
Enter the hostname for your local installation or for the CBIIT installation of caArray. If you misspell it, you will receive an error message.
caArray Experiment ID
Enter the caArray Experiment ID which you know corresponds with the copy number data.
Loading Type
Enter the Loading Type of the data file you plan to map.
Subject and Sample Mapping File
Browse for the appropriate CN mapping file. The file must be a CSV file with 3 column format for mapping data files (format: subject id, sample id, file name). Supplemental data uses 6 column-files.
Bioconductor Service Type
This is the type of bioconductor module that will be used for segmentation. Select between the two options: DNAcopy or CGHcall.
caCGHcallcaCGHcall call Service URL
Enter the URL for the grid segmentation service used to access the caCGHcall service. For more information, see caCGHCall
Call Level
An input parameter to CGHcall. This is the number of discrete values used to represent the copy number level. Select between two options: 3 (consisting of discrete values of -1, 0, 1) or 4 (consisting of discrete values -1, 0, 1, 2)
caDNACopycaDNACopy caDNACopy Service URL*
Control for selecting the URL which hosts the caDNACopy grid service. For more information, see
Change Point Significance Level
Significance levels for the test to accept change-points
Early Stopping Criterion
The sequential boundary used to stop and declare a change
Permutation Replicates
The number of permutations used for p-value computation
Random Number Seed
The segmentation procedure uses a permutation reference distribution. This should be used if you plan to reproduce the results.
- Click Save Segmentation Data Calculation Configuration for a genomic data source. On the screen upload a copy number mapping file and configure the parameters to be sent when computing segmentation data.
Note title Be Careful After a study has been deployed and the genomic source has been loaded, you cannot change these copy number parameters without reloading the data from caArray first.
Remapping Copy Number Data in a Deployed Study
Occasionally you may need to remap copy number data in a deployed study. To do so, follow these steps:
- Select the Manage Studies button and select Edit for the study you wish to remap.
- On the Edit Study page, select Edit under the Genomic Data Sources header.
- Without altering any information, select Save. When the warning box appears, select OK.
- Select ConfigureCopyNumberData.
- Enter the new mapping file in the Subject and Sample Mapping File field.
- Select Save Segmentation Data Calculation configuration.
- Select Deploy Study.
...
- On the Edit Study page under the Imaging Data Sources section, click the Add New button.
Info title Note If you have already provided an imaging data source, it is listed in this section of the Edit Study page. To edit the imaging data source, click the Edit button which opens the same dialog box described in the following steps.
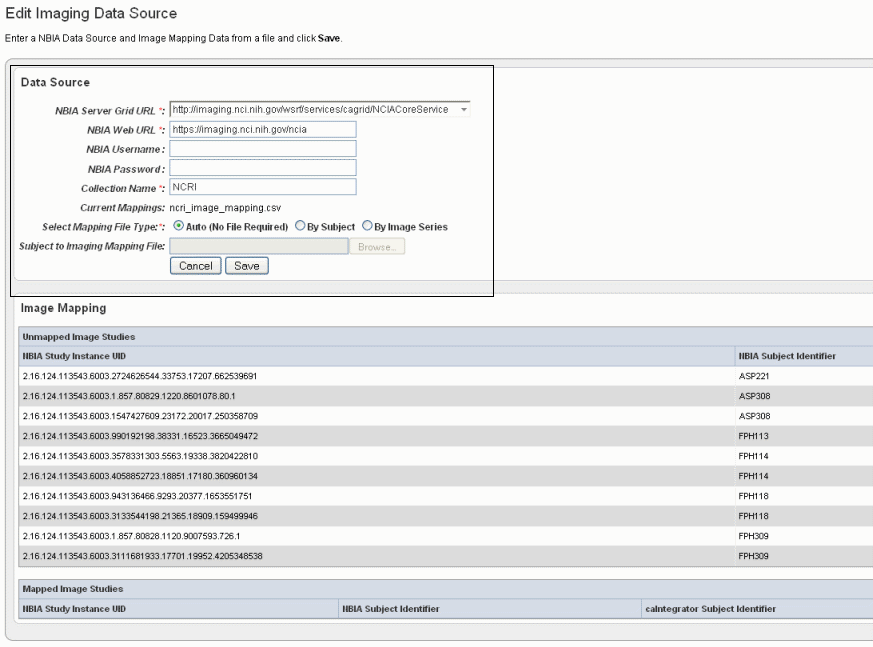
- In the Edit Imaging Data Source dialog box, configure the appropriate imaging data source information in the fields as shown in the selected area of the following figure and described below. Asterisks indicate required fieldsFields with an asterisk are required.
Fields
Description
NBIA Server Grid URL*
Enter the URL for the grid connection to NBIA.
NBIA Web URL
Enter the URL of the web interface of the NBIA installation.
NBIA Username and NBIA Password
This information is not required, as currently all data in the NBIA grid is Public data.
Collection Name
Enter the name/source for the collection you want to retrieve.
Current Mapping
If a mapping file has already been uploaded to the study to map imaging data, the file name displays here.
Select Mapping File Type
Click to select the file type:
--Auto – No file is required. Selecting this takes all subject annotation subject IDs and attempts to map them to the corresponding ID in the collection in NBIA. If the ID does not exist in NBIA, then no mapping is made for that ID.
--By Subject – Requires a mapping file to be uploaded. The "subject annotation to imaging mapping file" must be in CSV format with two columns that map the caIntegrator subject annotation subject ID to the NBIA subject ID.
--By Image Series – Requires a file to be uploaded. The subject annotation to imaging mapping file needs to be a two column mapping (CSV) from the caIntegrator subject annotation subject ID to the NBIA study instance UID.Subject to Imaging Mapping File
Click Browse to navigate to the appropriate subject annotation to imaging mapping file. See the Select Mapping File Type* field description.
Info title Note If mapping files have already been uploaded for the data sources you are editing, the Image Mapping tables of the dialog box show the mapping from NBIA Image Series Identifier to caIntegrator Subject Identifier.
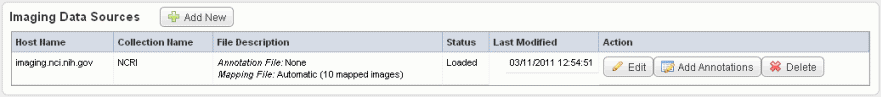
- Click Save to upload the data from NBIA to caIntegrator. The imaging data displays on the Edit Study page under the Imaging Data Sources section, as shown in the following figure.
- Once the data is uploaded, you can add image annotations. For more information, see #Adding or Editing Image Annotations.
...
Using either method, the image annotations are uploaded to caIntegrator. After this occurs, when you click the Edit Annotations button, the system opens to the Define Fields for Imaging Data page where you can edit the annotations. This is the same page (with a customized title) as that described in #Define Fields Page for Editing Annotations. You must assign identifiers and annotations to the data in the same way you did with the subject annotation data. For more information, see #Assigning an Identifier or Annotation and #Searching for Annotation Definitions.
Adding External Links
This If you are a study manager, this feature on the Edit Study page allows you to configure a CSV file with URLs to be used as external links relevant to a the study. This allows you to easily share or configure references.
To add an external link, follow these steps:
...
- On the Edit Study page, click the Add button under External Links section. External links can be any URL(s) to resources that are hosted external to caIntegrator but are relevant to the study being deployed.
- Assign a name to the external link.
- Add a description for the link, if appropriate.
- Browse for the CSV file containing URLs (HTTP linked) to resources outside of caIntegrator.
- Click Upload Now. caIntegrator does not validate any links in the file being uploaded.
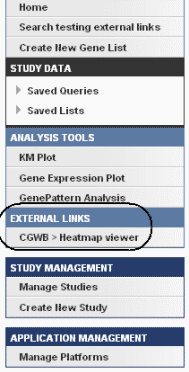
Once you have created external links for a study, when the study is open, an External Links section showing the link(s) displays on the left sidebar of the page , shows the link(s). An example is identified in the following figure.
Click the an external link to open a page that displays appropriately formatted web page links; an example is shown in the following figure.
Deploying the Study
...
Once you have started to create a study or have deployed it, you can update an existing the study in the following ways:
- Add new attributes (annotations) and upload relevant data to an the existing study.
- Delete a the study
- Modify existing annotation definitions
- Reload a subset of a study data and re-deploy the study and perform new analyses
- Re-deploy the entire study with new set of data and mappings.
...