|
Page History
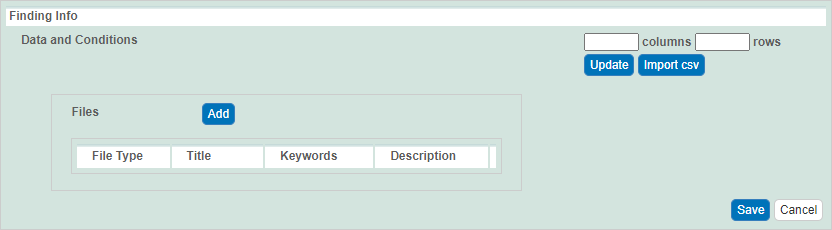
You can add purification data findings to a sample. (TBD: You can add as many as you wish?)
To add each finding:
On the Add or Edit page for synthesis purification of a sample, expand the Finding panel, as described in Adding or Editing Synthesis Purification in a Sample
.Add
Under Data and Conditions, addfindings such as laboratory conditions, pH, or temperature.
You can add data manually or you can import as many .
csvCSV files as needed.
(TBD: May I have a sample CSV file to import, to test these instructions?)To import a file of data values
Save a spreadsheet of data values to a csv CSV (comma-separated value) file.
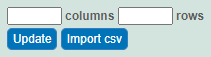
Click Import csvCSV. Select the data file and follow the prompts to add it. The system adds columns and data under Data and Conditions.
To add the data values manually
Specify the number of columns and rows for the matrix, and click Update.
Add the data values to the rows.
Whether you imported values or added values manually, you can preface each data value with one of the following: Maintain the default, equal to (=), or select greater than (>), less than (<), or infinity (approximate). (TBD: On the Dev ISB Test tier, these drop-down lists are empty.)
To define
aeach column,
clickperform the following steps:
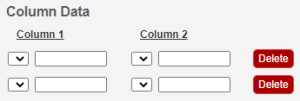
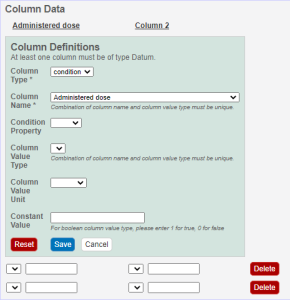
Click an underlined column heading. The Column Definition panel appears.
Select a Column Type, Datum or Condition.
Select a Column Name or select other and add a new one. You can add up to three cell viability Column Names, including cell viability, cell viability B, and cell viability C.
For Column Type Datum, (Column Name
options TBD: I need help with this.).
For Column Type Condition, (Column Name
options TBD:
I need help with this.).
To further identify a column, select a Column Value Type.
Once the column information is saved, the Column Value Type is shown in parentheses after the Column Name, such as cell viability (mean).
If necessary, select a
Column Value Unit, or select other and add one.
If you want the same value to fill all rows in a column, add a Constant Value. For Column Value Type, boolean, enter a Constant Value of 1 for true and 0 for false.
In the Column Definition panel, click Save. The system updates the columns.
If needed, click Set Column Order to change the order of the column headings in the matrix.
- Click Save in
In the Finding panel, click Save.