|
Page History
...
Topics in this chapter include the following:
| Table of Contents | ||
|---|---|---|
|
Query Results Overview
After you launch a search of a caIntegrator study, the system automatically opens the Query Results tab showing the results of your search. If you have not configured the column and sort display parameters before launching the search, by default the tab shows only the subject identifiers and a column that allows you to select each row of the data subset.
To display and/or sort additional data, you must return to the Results Type Tab and/or Sorting Tab to set display parameters, then re-run the search. The new search results will display the additional information, with the columns and data sorted as you specified.
...
You can download search results as a CSV file. The file contains the annotations, columns and data sort configurations you specified in the search query. See #Exporting Exporting Data.
See #Subject Subject Annotation and Imaging Data Results, #Gene Gene Expression Data Results, and #Expanding Expanding Imaging Data Results.
Subject Annotation and Imaging Data Results
...
If you run the search before configuring column and sort display parameters, only the \ [subject\] ID that meet the criteria and a column allowing you to select each row appear on the table, as shown in the following figure.
!imaging subj ID only75.png|vspace=4, alt="Query Results page, described in text."!figure.
You can add details for one or more subjects by configuring them on the Results Type tab. Annotations listed there are the column headers in the CSV file(s) that were uploaded to the study. For information about using the Results Type tab, see Results Type Tab.
...
For Gene criteria, caIntegrator finds and summarizes at the gene level all reporters that match criteria for the gene you defined on the Criteria tab. When displaying results at the gene level, the mean is displayed if there are two probes, the median if there is an odd number of probes greater than two and the mean of the two median probes if there is an even number of probes greater than two. Next to each gene symbol, caIntegrator displays an icon () which you can click to open the Cancer Genome Anatomy Project (CGAP) showing data for the gene. Gene symbols and icons are identified in the following figure.
If you have selected Gene Expression on the Results Type tab, then the column headers are a clickable label which sorts the entire table on that column. If you selected Reporter ID on the Results Type tab, the Reporter ID is clickable (and the gene is not clickable).
...
Gene reporter type display
Reporter ID reporter type display
...
You can save genes identified in the search results as a gene list. For more information, see #Creating Creating a Gene or Subject List.
Copy Number Data Results
After defining copy number criteria on the Criteria tab and running a copy number query, you should select the Copy Number result type on the Results Type Tab, and rerun the query. Copy number data search results display in a data matrix containing samples vs. genomic regions.
- Gene symbols display parallel to chromosome regions on the matrix.
- Sample ID column headings display the Subject ID/Sample ID (for example, E09262/E09262) because each calculation is based on a comparison of a tumor and matched blood sample from the same subject.
The values in the Sample ID columns are mean segment values as calculated by the DNAcopy algorithm, shown in the following figure. These are expressed as log2 (test/reference, as in tumor/normal). For more information about the algorithm, see Bioconductor
.
.Multiexcerpt include nopanel true MultiExcerptName ExitDisclaimer PageWithExcerpt wikicontent:Exit Disclaimer to Include DNAcopy ouput values can be negative. If the test and the reference genomic samples both have two copies of a chromosomal region, the ratio of test/reference is '1', and the log2(1) = 0. That is, if there is no change in the chromosomal structure, then the value is 0. If there are more copies in the test sample (amplification of the chromosomal segment), the ratio of test to reference is greater than 1, and the log2(test/reference) is greater than 0. For example, if the test sample has 6, the ratio or test/reference is 6/2 = 3; log2(3) = 1.58. In a deletion, the test is less than the reference, for example 1. The DNAcopy output value would be log2(1/2) = log2(0.5) = -1.0. Values below -0.6 are often considered a deletion.
...
- Click the Create New List link in the left sidebar. This opens the Manage List page, shown in the following figure.
- Enter a name for the gene or subject list.
- Enter a description (optional).
- Select Make Visible to Others if you want the list to be visible to anyone who views the study. This selection places the list in the Global List folder in the left sidebar under Saved Lists. In any box where you can select lists, the term 'Global' will identify any list so identified when the list is created.
- Select the List Type, Subject or Gene.
- If you select Subject, enter the Subject IDs in the text box that appears. Proceed with step 7.
- If you select Gene, proceed with step 6.
- For Gene Symbol, caIntegrator provides three methods whereby you can obtain gene symbols for creating a gene list. For more information, see #Choosing Choosing Genes.
- If you so choose, you can upload a gene or subject ID list. For the Upload File field, click the Browse button to navigate to a .csv file made up of gene symbols. caIntegrator converts the comma-separated content to a gene list.unmigrated-wiki-markup
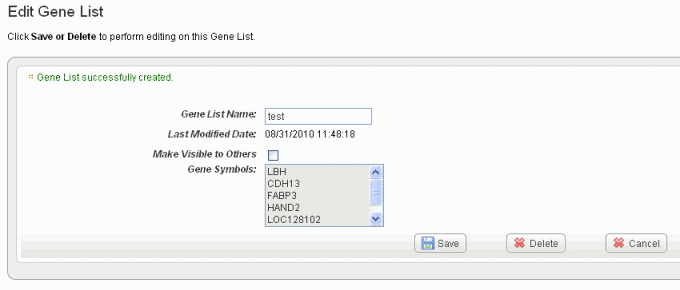
- Click *Create List* at the bottom of the page. caIntegrator now opens the Edit \ [Subject or Gene\] List page which shows the name and symbols of the newest gene list, shown in the following figure.
!edit gene list80.png|vspace=4, border=1, alt="The Edit Gene List for reviewing, editing the name, deleting a gene list and setting visibility for the list. The Edit Subject List page is comparable."!newest gene list, shown in the following figure.
See Editing See #Editing a Gene or Subject List for information about the edit feature.
| Info | ||
|---|---|---|
| ||
When you perform a GISTIC analysis, caIntegrator automatically saves the retrieved genes in the Saved Copy Number analysis in the left sidebar. For a query or plot analysis, they also appear in the Gene Picker dialog box described in #Choosing Choosing Genes. |
Editing a Gene or Subject List
...
- Click on any of the list names or the list icon () to rerun the query from which the gene or subject list was first created. If the list is a gene list, in the query results, you can click on the gene information icon () to open the Cancer Genome Anatomy Project (CGAP) showing metadata for the gene.
- Click the edit icon () to open an Edit Gene/Subject List dialog box. On this page you can review the list of gene symbols or subject IDs included in the list shown in the following figure.
unmigrated-wiki-markup - In the Edit \ [List Type\] dialog box, you can perform the following tasks:
- unmigrated-wiki-markup
- To rename the list in the *\[List Type\] List Name* text box, enter the new list name.
- You can change the visibility of the list in the appropriate check box.
- To delete the list, click the Delete button.
- Click Save to save your changes or Cancel to leave the page without making changes.
Once a list is created, you cannot edit the list contents.
| Include Page |
|---|
...
|
...
|
| Include Page |
|---|
...
|
...
|
Expanding Imaging Data Results
In reviewing imaging search results, it is important to understand the hierarchy of submissions in NBIA. For more information, see #Relationship Relationship of Subject to Study to Series to Images.
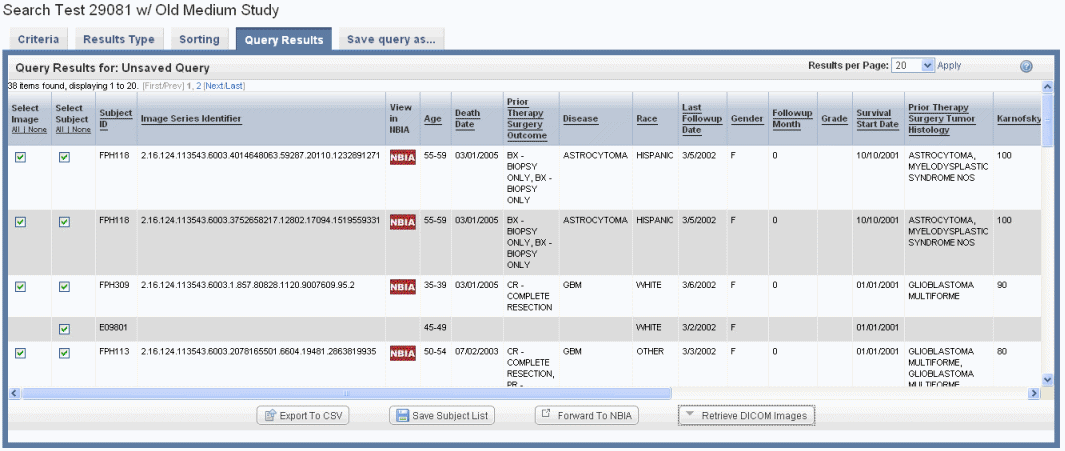
If you run a search before configuring column and sort display parameters, only the Subject Identifiers for the patients/images that meet the criteria and a column containing one check box per row display by default. An example displays in the following figure.
If your annotation choice on the Results Type page identifies annotations such as tumor size or tumor location, the search results display image series subsets that have those annotations, or any annotations you check on the Results Type page. The check boxes work in conjunction with buttons at the bottom of the results page, shown in the following figure. By expanding display parameters, you can view complete details for image search results.
You can add more details for images by configuring image annotations on the Results Type tab. Annotations listed there are the column headers in the image series CSV file(s) that were uploaded to the study. Examples of image details include the following:
...
You can set display parameters for the results on the Columns and Sorting tabs. For more information, see #Results Results Type Tab.
See also #caIntegrator caIntegrator and NBIA, [#Retrieving Retrieving DICOM Images and #Example Example of Retrieving Images.
caIntegrator and NBIA
Images can be accessed in NBIA if you see buttons on the Search Results page. See the Imaging Note in #Results Results Type Tab. You can click links on the Search Results tab to view or download image data.
...
On the caIntegrator imaging data Search Results page, you can click the Retrieve DICOM Images button which is linked to results you have selected by row. caIntegrator retrieves the corresponding image(s) from NBIA through the grid. NBIA organizes the download file by patient ID, StudyInstance UID, and ImageSeries UID, and compresses it into a zip file. When caIntegrator notifies you that the file is retrieved, the DICOM Retrieval page indicates whether the retrieved files are Study Instance UIDs or Image Series UIDs, shown in the following figure.
Click the Download DICOM link to download and save the file. caIntegrator unzips the file and displays the list of images in the file. To open the DICOM images, you must have a DICOM image viewer application installed on your computer. For more information about one such viewer, see http://www.e-dicom.com/viewers.php
| Multiexcerpt include | ||||||
|---|---|---|---|---|---|---|
|
In the search results, not all of the subjects in the data subset may be mapped to image series IDs. If you select a mixture of subjects, some of which have image annotations as indicated by an image series ID and some of which do not have image annotations (no image series ID), when you click the Retrieve DICOM Images button, NBIA retrieves the images for the entire NBIA study instance UID that includes the image seriesIDs you checked.
...
To best understand this, it is important to review the hierarchy of submissions in NBIA. For more information, see #Relationship Relationship of Subject to Study to Series to Images.
...
When the image Study is in the checked boxes (regardless of image series being there or not), the system aggregates up to the Image Study level.
| Include Page |
|---|
...
|
...
|
Exporting Data
You can choose to download tabular search results as a CSV file. Click the Export .csv link at the bottom of the page. You may need to scroll the page to see it. The file contains the annotations, columns and data sort configurations you specified in the search query.
...