|
Page History
...
- You can specify visibility of specified annotation data in the Visiblecolumn.
- Select a checkbox for a row to make the corresponding data visible to all subscribers of the study or anonymous users if the study is made available to the public.
- Clear a checkbox to hide the corresponding annotation from any subscriber or anonymous user of the study. Data continues to exist but does not show up in query fields nor in query results.
- The Annotation Header from File column on the Define Fields for Subject (or Image) Data page (the figure shown above) displays column headers taken from the source CSV file. The Define Fields... page also displays data values in the file you have designated. You must map each column name to an existing column name in the caIntegrator database or in caDSR. If it doesn't yet exist, you can create a custom column name. The following figure is an example CSV file whose data you would be mapping in caIntegrator in the Define Fields page.
- To indicate the unique identifier of choice, on the row showing the column header (PatientID in the figure, but other examples are subject identifier, sample identifier, etc), click Change Assignment in the Annotation Definition column.
...
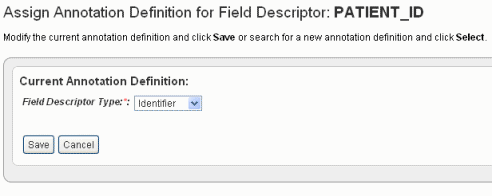
- For the column that you choose to be the one and only Identifier column (in this case, PatientID), in the Column Type drop-down list, select Identifier. The following figure shows the dialog box rendering when "identifier" is selected in the Field Descriptor Type drop-down list.
- Click Save to save the identifier. This returns you to the Define Fields for Subject Data page where the Identifier is noted in the Annotation Definition column.
- After you have defined which field is the Identifier, you must ensure that ALL other data fields also have an annotation definition assignment. For those fields without an annotation definition assignment or for those whose annotation definition you want to review, click Change Assignment.
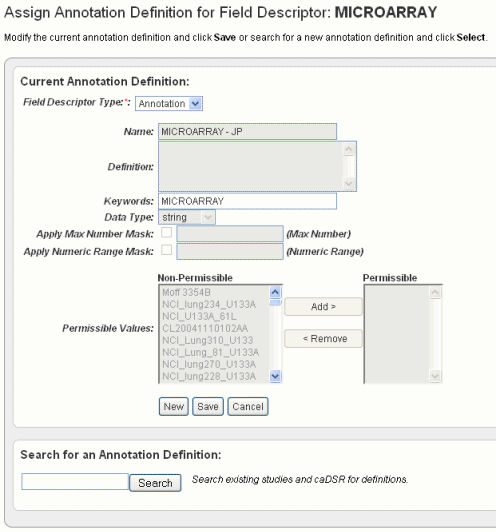
- In the Assign Annotation Definition for Field Descriptor dialog box, shown in the following figure, select Annotation in the drop-down list.
As you select the column type, you can work with column headers in one of four ways in this dialog box.- You can accept existing default definitions (those that are inherent in the data file you selected). See Step 5.
- You can create and/or manage your own definitions manually. See Step 6.
- You can search for and use definitions in other caIntegrator studies. See Searching for Annotation Definitions.
- You can search for and use definitions found in caDSR. See Searching for Annotation Definitions.
- Review the current annotation definition in the Assign Definition page, Current Annotation Definition section. Click Cancel to return to the Define Fields... page.
You can still initiate a search for another annotation definition in the Search for an Annotation Definition section on the browser page if you choose to change the definition. See the bottom section of the preceding figure. See also Searching for Annotation Definitions. Click Save to retain any changes. To enter a new name annotation, or any other information about the annotation definition, click the Newbutton and enter the information described in the following table.
Annotation Field
Field Description
Name
Enter the name for the annotation.
Definition
Enter the term(s) that define the annotation.
Keywords
Insert keyword(s) that could be used to find the annotation in a search, separated by commas.
Data Type
Select a string (default), numeric, or date.
Apply Max Number Mask
This field is available only for numeric-type annotations, or when a new definition is created. This feature is unavailable when permissible values are present.
Select the box and enter a maximum number for the mask, such as "80" for age. When you query results above the value of the mask, then the system displays the mask and not the actual age.Tip title Tip If you enter masks of both "max number" and "range", caIntegrator applies both masks at the same time.
The Data Dictionary page now has a Restrictions column that shows restrictions whenever a mask has been applied.Apply Numeric Range Mask
This field is available only for numeric-type annotations, or when a new definition is created. This feature is unavailable when permissible values are present.
Select the box and enter a width of range for the mask, such as "5" representing blocks of 5 years. For example, if you enter a width of 5, the query only allows age blocks of 0-5, 6-10, 11-15, etc.
When you query results above the value of the mask, then the system displays the mask and not the actual age ranges.Tip title Tip If you enter masks of both "max number" and "range", caIntegrator applies both masks at the same time.
The Data Dictionary page now has a Restrictions column that shows restrictions whenever a mask has been applied.Permissible/Non-permissible Values
Tip title Tip The first time you load a file, before you assign annotation definitions, step #3 in Assigning an Identifier or Annotation, these panels may be blank. If the column header for the data is already "recognizable" by caIntegrator, the system makes a "guess" about the data type and assigns the values to the data type in the newly uploaded file. They will display in the Non-permissible values sections initially. Use the Add and Remove buttons to move the values shown from one list to the other, as appropriate.
...
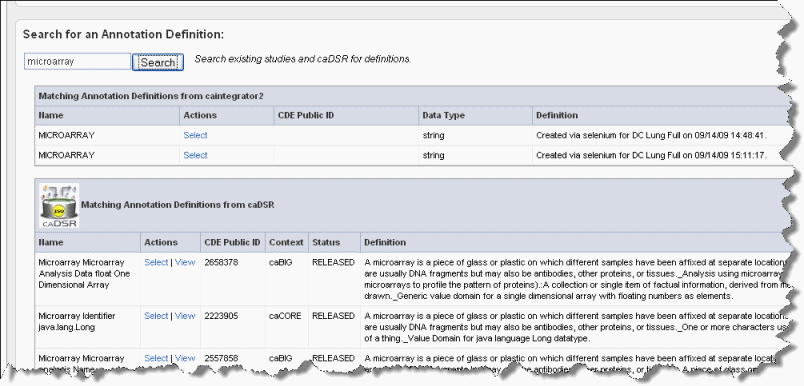
- Enter search keyword(s) in the Search text box on the Assign Annotation Definition page (the preceding figure). Click Search or click Enter to launch the search. After a few moments, the search results display on the same page. An example of search results is shown in the following figure.
To view the definitions corresponding to any of the "Matching Annotation Definitions", which are those currently found in other caIntegrator studies, click the [term], such as "age", hypertext link. The definition then appears in the Current Annotation Definition segment of the page just above.
Info title In Summary When you click the link for a definition, that assigns the definition to the Define Fields for Subject Data page, and it also closes the Annotation Definition page. You can modify any portion of the definition, as described in Step 6 in Assigning an Identifier or Annotation.
The matches from caDSR display some of the details of the search results. To view more details of a match, such as permissible values, click View, which opens caDSR to the term. If you click Select, the caDSR definition automatically replaces the annotation definition for this field with which you are working.
Note title Caution Take care before you add a caDSR definition that it says exactly what you want. caDSR definitions can have minor nuances that require specific and limited applications of their use.
Once you have settled on an appropriate field definition for the annotation, click Save. This returns you to the Define Fields for Subject Datapage.
Info title Note If you have not clicked Select for alternate definitions in this dialog box, then click Save to return to the Define Field...dialog box without making any definition changes.
From the Define Fields for Subject Data page, be sure and designate the data types for each field in the file. Click Save on each page to save your entries or click New to clear the fields and start again. You will not be able to proceed until every field definition entry on the Fields for Subject Data screen has an entry, one as the unique Identifier and the remainder as annotations.
The Data From File columns on the Define Fields... page display the column header values of the first three rows you designated as "annotations".Tip title Tip Saving your entries in this way saves the study by name and description, but does not deploy the study. See Deploying the Study.
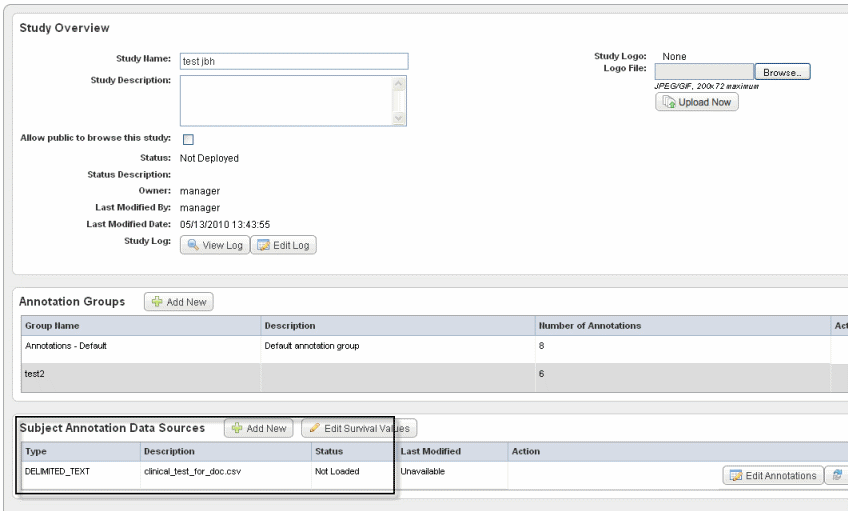
The Edit Study page now displays a "Not Loaded" status for the file whose annotations (column headers) you have defined. An example of a file whose annotations have been defined but not yet loaded is shown in the following figure.
Status definitions:- Definition Incomplete – An annotation definition or definitions must be modified on the Define Fields for Subject Data page. This status may be displayed because an identifier has not been selected. See Define Fields Page for Editing Annotations.
- Not Loaded – The annotation definitions must be loaded before a study can be deployed. If an error appears after attempting to load a subject annotation source, cick the Edit Annotations button which takes you to the Define Fields for Subject Data page where the problematic annotations will appear in red. See Define Fields Page for Editing Annotations.
- Loaded – The annotation definitions are properly loaded.
Click the Load Subject Annotation Source button in the Action section to load the data file you have configured, The Deploy Studybutton, to this point has been unavailable, but this step activates the button.
Tip title Adding Files You can add as many files as are necessary for a study. Patients 1-20 in first file, 21-40 in second file, or many patients in first file and annotations in second file, etc. As long as IDs are defined correctly, it works.
Click Deploy Study. caIntegrator now loads data from the file to the caIntegrator database, and the file status changes to "Loaded".
Tip title Changing Assignments You can change assignments even after the study is deployed, using the Edit feature. For more information, see Creating or Editing a Study.
...
caIntegrator goes to caArray, validates the information you have entered here, finds the experiment and retrieves all the sample IDs in the experiment. Once this finishes, the experiment information displays on the caIntegrator Edit Study page under the Genomic Data Sources section, as shown in the following figure. caIntegrator refreshes carray data on an hourly basis. The status of caArray data updates displays on the Manage Studies page and on the Edit Sample Mappings page.
| Info | ||
|---|---|---|
| ||
If you want to redefine the caArray experiment information, you can edit it. Click the Edit link corresponding to the Experiment ID. The Edit Genomic Data Source dialog box reopens, allowing you to edit the information. |
...
- Start with the 6-column mapping file template, described as follows:
- All platforms – Raw (level 1) data cannot be mapped; only normalized, processed (level 2) data is acceptable.
- The required six-column file format uses the following columns:
- Subject ID
- Sample ID
- Name of supplemental file (if appropriate, as attached to the experiment in caArray)
- Probe Header – Name of column header (in the supplemental file) which contains the probe IDs.
- Value Header – Name of column header (in the supplemental file) which holds the level 2 data.
Sample Header– Name of column header (in the supplemental file) which holds the level 2 data.
Info title Last two columns Only one of the last 2 columns is used: a single sample per file uses the Value Header column; multiple samples per file used Sample Header column. Unused columns are blank.
The following figure shows an example multiple sample mapping file in CSV format.
- When you use the mapping file, make sure you use the subject ID for mapping. If the file is human data, the subject ID is the patient ID.
- Determine whether your data in caArray is "imported and parsed" or "supplemental". These are the 'Loading Types' referred to in Step 4 of Steps for Mapping Genomic Data. Fill in the 6-column mapping file according to the following standard:
- Imported and parsed – Complete only the first two columns of the 6-column mapping file as described above. You can ignore the remaining columns.
Supplemental– Supplemental data comes in two types: "single sample per file" and "multiple samples per file". In either case, only one of the last two columns is used. If the supplemental data format is, single sample per file, the column named "Sample_Header" can be left empty. If the supplemental data format is multiple samples per file, the column named "Value_Header" can be left empty.
Info title Configuring supplemental files Supplemental files from caArray for mapping data must be configured appropriately. For information, see Supplemental Files Configuration.
The following steps use data of either type.
...
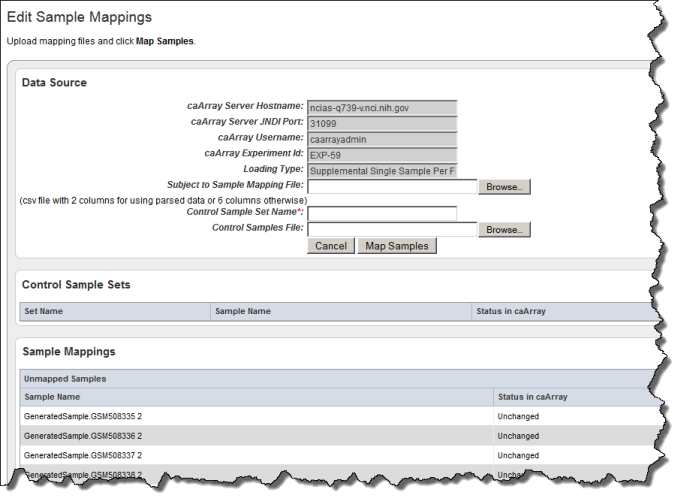
- On the Edit Study page, click the Map Samples button. This opens the Edit Sample Mappings page, shown in the following figure.
The first two caArray fields may be populated with the information for the instance of caArray to which you have access. You can, however, enter the caArray information described in the following table, if you prefer.
Field
Description
caArray Host Name
Enter the hostname for your local installation or for the CBIIT installation of caArray. If you misspell it, you will receive an error message.
caArray JNDI Port
Enter the appropriate server port. See your administrator for more information. Example: For the CBIIT installation of caArray, enter 8080 .
caArray Username
Enter your caArray account user name and password; you must have permissions in caArray for the experiment if it is private. If the data is public, you can leave this field blank.
caArray Experiment ID
Enter the caArray Experiment ID which you know corresponds with the subject annotation data you uploaded. Example: Public experiment "beer-00196" on the CBIIT installation of caArray (array.nci.nih.gov). If you misspell your entry, you will receive an error message.
- Enter the Loading Type of the data file you plan to map. (File types are described in Creating a Mapping File).
- In the Subject to Sample Mapping File section, click Browse to navigate for the Sample Mapping CSV file that you created (described in Creating a Mapping File). This provides caIntegrator with the information for mapping patients to caArray samples.
- Click the Map Samples button.
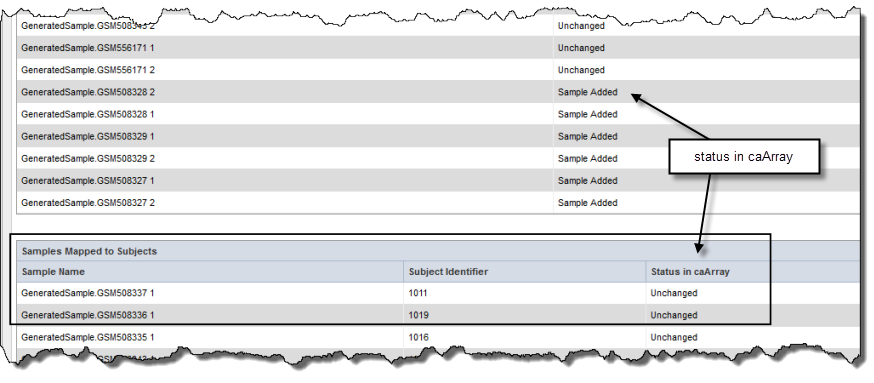
If the caArray data you have identified is imported and parsed, when you click the Map Samples button, the mapping takes place as the data is uploaded into caIntegrator. If the caArray data is supplemental, the mapping does not occur until the study is deployed. Mapped samples are listed in the Samples Mapped to Subjects section; scroll down the page to view them (see the following figure). caIntegrator refreshes caArray data hourly; once samples are mapped, a column on this page displays the status of the data in caArray (noted in the figure). Unmapped samples show at the top of the caIntegrator page. They were loaded from caArray, but they are not in the mapping file. These are not used for integration.
Info title Mapped or unmapped? If you have already mapped samples, when you first open this page they are listed in the Samples Mapped to Subjects section. If you have not already mapped samples, all of the samples in the caArray experiment you selected are listed as unmapped, because caIntegrator does not know how these sample names correlate to the patient data in the subject annotation file until you upload the subject to sample mapping file.
...
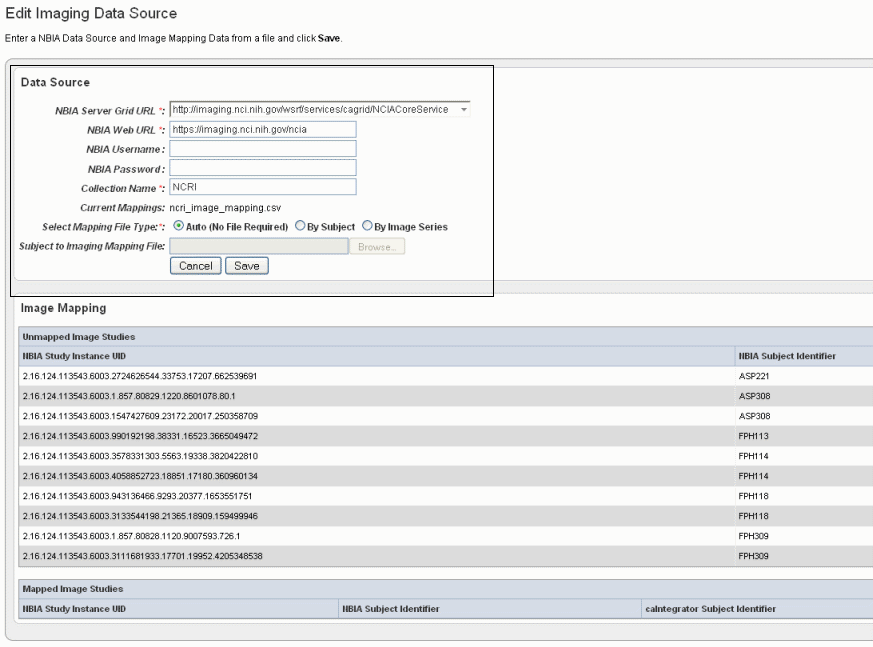
On the Edit Study page under the Imaging Data Sources section, click the Add Newbutton.
Info title Imaging data source If you have already provided an imaging data source, it is listed in this section of the Edit Study page. To edit the imaging data source, click the Edit button which opens the same dialog box described in the following steps.
In the Edit Imaging Data Source dialog box, configure the appropriate imaging data source information in the fields as shown in the selected area of the following figure and described below. Fields with an asterisk are required.
Field
Description
NBIA Server Grid URL*
Enter the URL for the grid connection to NBIA.
NBIA Web URL
Enter the URL of the web interface of the NBIA installation.
NBIA Username and NBIA Password
This information is not required, as currently all data in the NBIA grid is Public data.
Collection Name
Enter the name/source for the collection you want to retrieve.
Current Mapping
If a mapping file has already been uploaded to the study to map imaging data, the file name displays here.
Select Mapping File Type
Click to select the file type:
--Auto – No file is required. Selecting this takes all subject annotation subject IDs and attempts to map them to the corresponding ID in the collection in NBIA. If the ID does not exist in NBIA, then no mapping is made for that ID.
--By Subject – Requires a mapping file to be uploaded. The "subject annotation to imaging mapping file" must be in CSV format with two columns that map the caIntegrator subject annotation subject ID to the NBIA subject ID.
--By Image Series – Requires a file to be uploaded. The subject annotation to imaging mapping file needs to be a two column mapping (CSV) from the caIntegrator subject annotation subject ID to the NBIA study instance UID.Subject to Imaging Mapping File
Click Browse to navigate to the appropriate subject annotation to imaging mapping file. See the Select Mapping File Type* field description.
Info title Mapping files uploaded? If mapping files have already been uploaded for the data sources you are editing, the Image Mapping tables of the dialog box show the mapping from NBIA Image Series Identifier to caIntegrator Subject Identifier.
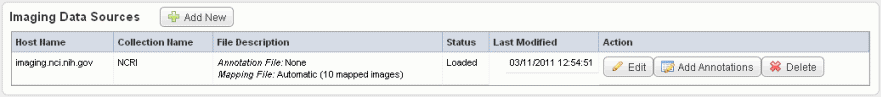
- Click Save to upload the data from NBIA to caIntegrator. The imaging data displays on the Edit Study page under the Imaging Data Sources section, as shown in the following figure.
- Once the data is uploaded, you can add image annotations. For more information, see Adding or Editing Image Annotations.
...
Click an external link to open a page that displays appropriately formatted web page links; an example is shown in the following figure.
Deploying the Study
...