|
Page History
...
The name specified in the third column of the mapping file is specific for each array manufacturer as follows:
- Affymetrix – The third column of the mapping file must contain filenames that end in .cnchp. The corresponding experiment in caArray must have these files and the extensions must match .cnchp.
- Agilent – The third column must name a file which contains level 2 copy number data. Level one copy number will not work. This file name is repeated for each line in the mapping file.
...
- In the Genomic Data Sources section of the Edit Study page, for the data you have already added, click Configure Copy Number Data button.
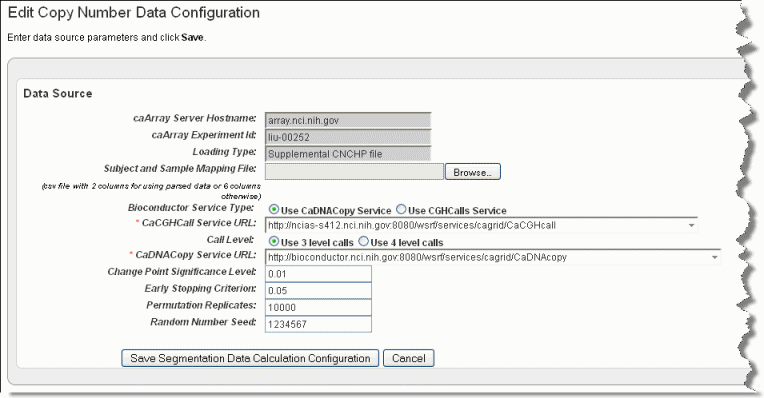
The Edit Copy Number page, shown in the following figure, opens.Info title Note This link is available only if you have uploaded copy number data and you are configuring a Copy Number data type (as indicated by the Data Type column on the Edit Study page).
- Browse for and enter appropriate information to identify and retrieve the copy number mapping file. The fields are described in the following table. An asterisk indicates a required field.
Field
Description
caArray Service Host Name
Enter the hostname for your local installation or for the CBIIT installation of caArray, . If you misspell it, you will receive an error message.
caArray Experiment ID
Enter the caArray Experiment ID which you know corresponds with the copy number data.
Loading Type
Enter the Loading Type of the data file you plan to map.
Subject and Sample Mapping File
Browse for the appropriate CN mapping file. The file must be a CSV file with 3 column format for mapping data files (format: subject id, sample id, file name). Supplemental data uses 6 column-files.
Bioconductor Service Type
This is the type of bioconductor module that will be used for segmentation. Select between the two options: DNAcopy or CGHcall.
caCGHcallcaCGHcall Service URL
Enter the URL for the grid segmentation service used to access the caCGHcall service. For more information, see caCGHCall
Call Level
An input parameter to CGHcall. This is the number of discrete values used to represent the copy number level. Select between two options: 3 (consisting of discrete values of -1, 0, 1) or 4 (consisting of discrete values -1, 0, 1, 2)
caDNACopycaDNACopy Service URL*
Control for selecting the URL which hosts the caDNACopy grid service For more information, see
Change Point Significance Level
Significance levels for the test to accept change-points
Early Stopping Criterion
The sequential boundary used to stop and declare a change
Permutation Replicates
The number of permutations used for p-value computation
Random Number Seed
The segmentation procedure uses a permutation reference distribution. This should be used if you plan to reproduce the results.
- Click Save Segmentation Data Calculation Configuration for a genomic data source. On the screen upload a copy number mapping file (format: subject id, sample id, file name) and configure the parameters to be sent when computing segmentation data.
Note title Be Careful After a study has been deployed and the genomic source has been loaded, you cannot change these copy number parameters without reloading the data from caArray first.
...
To add images from NBIA to the study you are creating, follow these steps:
- On the Edit Study page, under the Imaging Data Sources section, click the Add New button.
Info title Note If you have already provided an imaging data source, it is listed in this section of the Edit Study page. To edit the imaging data source, click the Edit button which opens the same dialog box described in the following steps.
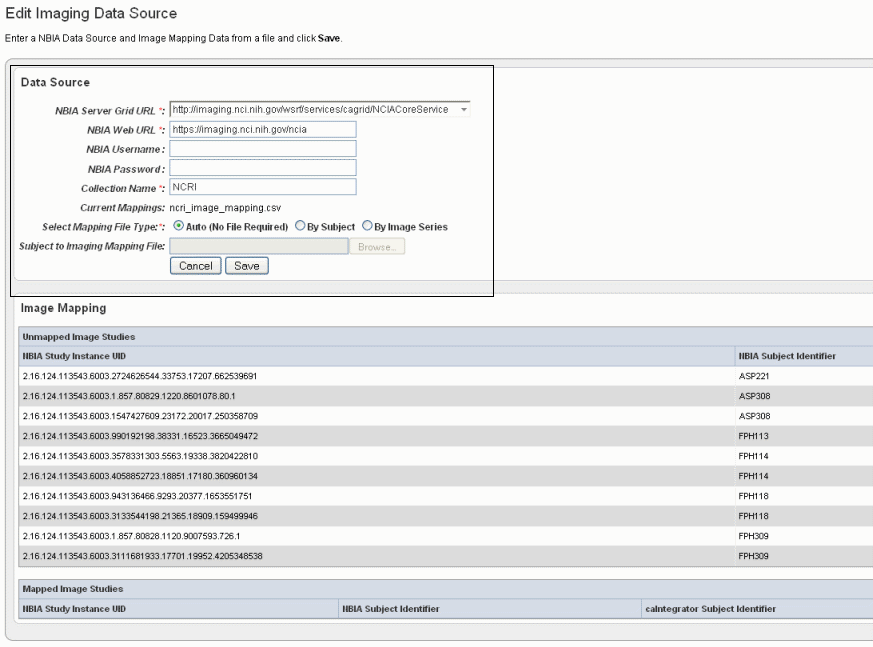
- In the Edit Imaging Data Source dialog box, configure the appropriate imaging data source information in the fields , as shown in the selected area of the following figure and described below. Asterisks indicate required fields.
–Fields
Description
NBIA Server Grid URL*
Enter the URL for the grid connection to NBIA.
- NBIA Web URL *– |Enter the URL of the web interface of the NBIA installation.| .
NBIA Username and NBIA Password
* –This information is not required, as currently all data in the NBIA grid is Public data.
Collection Name
–Enter the name/source for the collection you want to retrieve.
Current Mapping
* –If a mapping file has already been uploaded to the study to map imaging data, the file name displays here.
Select Mapping File Type*
Click to select the file type:
- Auto – No file is required. Selecting this takes all subject annotation subject IDs and attempts to map them to the corresponding ID in the collection in NBIA. If the ID does not exist in NBIA, then no mapping is made for that ID.
- By Subject – Requires a mapping file to be uploaded. The "subject annotation to imaging mapping file" must be in CSV format with two columns that map the caIntegrator subject annotation subject ID to the NBIA subject ID.
- By Image Series – Requires a file to be uploaded. The subject annotation to imaging mapping file needs to be a two column mapping (CSV) from the caIntegrator subject annotation subject ID to the NBIA study instance UID.| –
Subject to Imaging Mapping File
Click Browse to navigate to the appropriate subject annotation to imaging mapping file. See Select Mapping File Type* field description.
Info title Note If mapping files have already been uploaded for the data sources you are editing, the Image Mapping tables of the dialog box show the mapping from NBIA Image Series Identifier to caIntegrator Subject Identifier.
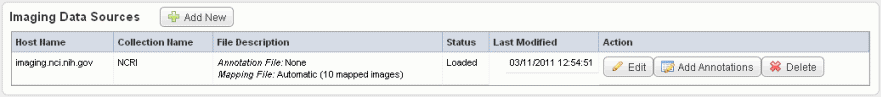
- Click Save to upload the data from NBIA to caIntegrator. The imaging data displays on the Edit Study page under the Imaging Data Sources section, as shown in the following figure.
- Once the data is uploaded, you can add image annotations. For more information, see #Adding or Editing Image Annotations.
...