|
Page History
...
- On the Edit Study page where you have selected and added the subject annotation data, click the Add New button under Genomic Data Sources. You can upload genomic data only from caArray.
This opens the Edit Genomic Data Source dialog box. Enter the appropriate information in the fields, shown in the following figure. The fields are described below.
Field Types
Field Description
caArray Web URL
Enter the URL for the caArray instance to be used for the genomic data sources. This will enable a user to link to the referenced caArray experiment from the study summary page.
caArray Host Name
Enter the hostname for your local installation or for the CBIIT installation of caArray, If you misspell it, you will receive an error message.
caArray JNDI Port
Enter the appropriate server port. See your administrator for more information. Example: For the CBIIT installation of caArray, enter 8080.
caArray Username and caArray Password
If the data is private, you must enter your caArray account user name and password; you must have permissions in caArray for the experiment. If the data is public, you can leave these fields blank.
caArray Experiment ID
Enter the caArray Experiment ID which you know corresponds with the subject annotation data you uploaded. Example: Public experiment "beer-00196" on the CBIIT installation of caArray (array.nci.nih.gov). If you misspell your entry, you will receive an error message.
Vendor
Select either Agilent or Affymetrix.
Data Type
Select Expression or Copy Number.
Platform
If appropriate, select the Agilent or Affymetrix platform.
Info title Note Because you can add more than one set of genomic data to a study, a study can also have multiple platforms, one for each set of genomic data.
Central Tendency for Technical Replicates
– If more than one hybridization is found for the reporter, the hybridizations will be represented by this method.
Indicate if technical replicates have high statistical variability
– If more than one hybridization is found, checking this box will display a ** in the genomic search results when a reporter value has high statistical variability.
Standard Deviation Type
- When the checkbox for indicating if technical replicates have high statistical variability is checked, this parameter becomes available. Select in the drop-down the calculation to be used to determine whether or not to display a ** (see previous bullet point).
--
- Relative{*}, which calculates the Relative Standard Deviation in percentage value
...
- Normal{*}, which calculates the Standard Deviation in numeric value|
Standard Deviation Threshold
When the checkbox for indicating if technical replicates have high statistical variability is checked, this parameter becomes available. This is the threshold at which the Standard Deviation Type is exceeded and the reporter is marked with a **.
- Click Save.
caIntegrator goes to caArray, validates the information you have entered here, finds the experiment and retrieves all the sample IDs in the experiment. Once this finishes, the experiment information displays on the caIntegrator Edit Study page under the Genomic Data Sources section, as shown in the following figure.
| Info | ||
|---|---|---|
| ||
If you want to redefine the caArray experiment information, you can edit it. Click the Edit link corresponding to the Experiment ID. The Edit Genomic Data Source dialog box reopens, allowing you to edit the information. |
Mapping Genomic Data to Subject Annotation Data
...
- Start with the 6-column mapping file template, described as follows:
- All platforms – Raw (level 1) data cannot be mapped; only normalized, processed (level 2) data is acceptable.
- The required six-column file format uses the following columns:
- Subject ID
- Sample ID
- Name of supplemental file (if appropriate, as attached to the experiment in caArray)
- Probe Header – Name of column header (in the supplemental file) which contains the probe IDs.
- Value Header – Name of column header (in the supplemental file) which holds the level 2 data.
- Sample Header – Name of column header (in the supplemental file) which holds the level 2 data.
Info title Note Only one of the last 2 columns is used: a single sample per file uses the Value Header column; multiple samples per file used Sample Header column. Unused columns are blank.
The following figure shows an example multiple sample mapping file in CSV format.
- When you use the mapping file, make sure you use the subject ID for mapping. If the file is human data, the subject ID is the patient ID.
- Determine whether your data in caArray is "imported and parsed" or "supplemental". These are the 'Loading Types' referred to in Step 4 of #Steps for Mapping Genomic Data. Fill in the 6-column mapping file according to the following standard:
- Imported and parsed – Complete only the first two columns of the 6-column mapping file as described above. You can ignore the remaining columns.
- Supplemental – Supplemental data comes in two flavors: "single sample per file" and "multiple samples per file". Only one of the last two columns is used. If the supplemental data format is:
- Single sample per file – the column named "Sample_Header" can be left empty.
- Multiple samples per file – the column named "Value_Header" can be left empty.
| Info | ||
|---|---|---|
| ||
Supplemental files from caArray for mapping data must be configured appropriately. For information, see Supplemental Files Configuration. |
The following steps use data of either type.
Steps for Mapping Genomic Data
...
- On the Edit Study page, click the Map Samples button. This opens the Edit Sample Mappings page, shown in the following figure.
- The first two caArray fields may be populated with the information for the instance of caArray to which you have access. You can, however, enter the following caArray information described in the following table, if appropriate. –
Field
Description
caArray Host Name
–Enter the hostname for your local installation or for the CBIIT installation of caArray, If you misspell it, you will receive an error message.
caArray JNDI Port
–Enter the appropriate server port. See your administrator for more information. Example: For the CBIIT installation of caArray, enter 8080.
caArray Username
–Enter your caArray account user name and password; you must have permissions in caArray for the experiment if it is private. If the data is public, you can leave this field blank.
caArray Experiment ID
Enter the caArray Experiment ID which you know corresponds with the subject annotation data you uploaded. Example: Public experiment "beer-00196" on the CBIIT installation of caArray (array.nci.nih.gov). If you misspell your entry, you will receive an error message.
- Enter the Loading Type of the data file you plan to map. (File types are described in #Creating a Mapping File).
- In the Subject to Sample Mapping File section, click Browse to navigate for the Sample Mapping CSV file that you created (described in #Creating a Mapping File). This provides caIntegrator with the information for mapping patients to caArray samples.
- Click the Map Samples button.
If the caArray data you have identified is imported and parsed, when you click the Map Samples button, the mapping takes place as the data is uploaded into caIntegrator. If the caArray data is supplemental, the mapping does not occur until the study is deployed. - Mapped samples will be listed in the Samples Mapped to Subjects section; scroll down the page to view them (see the following figure). Unmapped samples show at the top of the caIntegrator page. They were loaded from caArray, but they are not in the mapping file. These are not used for integration.
Info title Note If you have already mapped samples, when you first open this page they are listed in the Samples Mapped to Subjects section. If you have not already mapped samples, all of the samples in the caArray experiment you selected are listed as unmapped, because caIntegrator does not know how these sample names correlate to the patient data in the subject annotation file until you upload the subject to sample mapping file.
...
The name specified in the third column of the mapping file is specific for each array manufacturer as follows:
- Affymetrix – The third column of the mapping file must contain filenames that end in .cnchp. The corresponding experiment in caArray must have these files and the extensions must match .cnchp.
- Agilent – The third column must name a file which contains level 2 copy number data. Level one copy number will not work. This file name is repeated for each line in the mapping file.
...
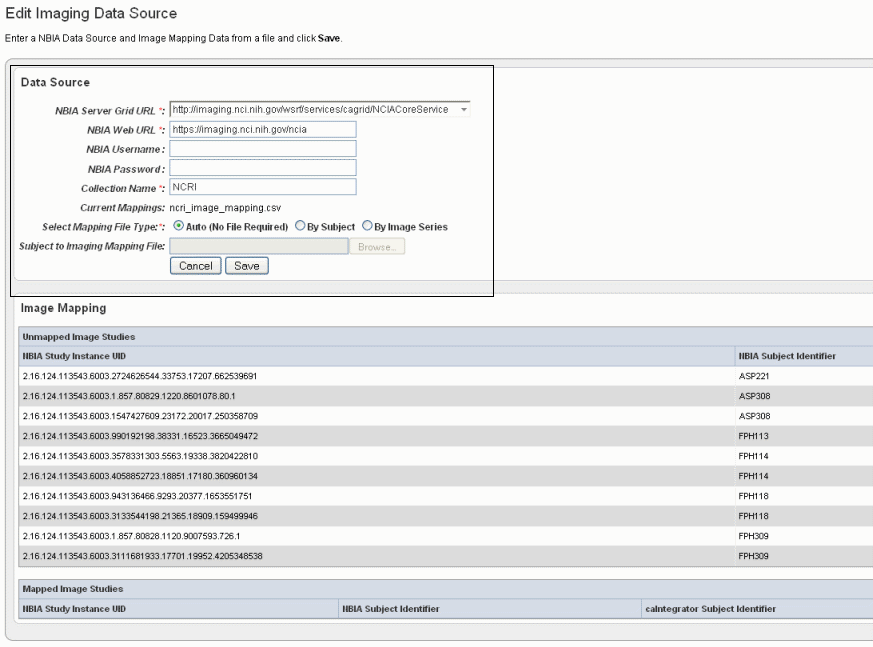
- On the Edit Study page under the Imaging Data Sources section, click the Add New button.
Info title Note If you have already provided an imaging data source, it is listed in this section of the Edit Study page. To edit the imaging data source, click the Edit button which opens the same dialog box described in the following steps.
- In the Edit Imaging Data Source dialog box, configure the appropriate imaging data source information in the fields as shown in the selected area of the following figure and described below. Asterisks indicate required fields.
Fields
Description
NBIA Server Grid URL*
Enter the URL for the grid connection to NBIA.
NBIA Web URL
...
Enter the URL of the web interface of the NBIA installation.
...
NBIA Username and NBIA Password
This information is not required, as currently all data in the NBIA grid is Public data.
Collection Name
Enter the name/source for the collection you want to retrieve.
Current Mapping
If a mapping file has already been uploaded to the study to map imaging data, the file name displays here.
Select Mapping File Type
...
Click to select the file type:
- Auto – No file is required. Selecting this takes all subject annotation subject IDs and attempts to map them to the corresponding ID in the collection in NBIA. If the ID does not exist in NBIA, then no mapping is made for that ID.
- By Subject – Requires a mapping file to be uploaded. The "subject annotation to imaging mapping file" must be in CSV format with two columns that map the caIntegrator subject annotation subject ID to the NBIA subject ID.
- By Image Series – Requires a file to be uploaded. The subject annotation to imaging mapping file needs to be a two column mapping (CSV) from the caIntegrator subject annotation subject ID to the NBIA study instance UID.|
Subject to Imaging Mapping File
Click Browse to navigate to the appropriate subject annotation to imaging mapping file. See the Select Mapping File Type* field description.
Info title Note If mapping files have already been uploaded for the data sources you are editing, the Image Mapping tables of the dialog box show the mapping from NBIA Image Series Identifier to caIntegrator Subject Identifier.
...