Topic: caIntegrator Usage
Release: caIntegrator 1.2
Date entered: August 31, 2010
Question
How to search for pathway in caIntegrator?
Answer
The user can launch the search for pathway in caBio within caIntegrator by following the steps below:
Step 1: Select a study.
The pathway search can be done in either a private or a public study (no login required). In the example below, we used the public study, hosted on caIntegrator's public domain.
Select a study from the drop-down list in the top tool bar (DC Lung Study in this example).
Step 2 : Initiate Search within the study.
The left menu bar shows the options available within the study. Click on Search DC Lung Study from the left menu bar.
Step 3 Initiate Search for the gene
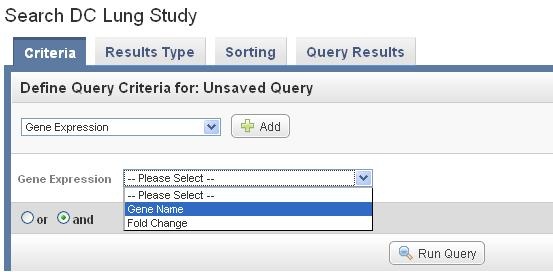
Select Gene expression under the Criteria tab in the main section of the study, and Click on Add next to the option. After the field of Gene Expression appears, select Gene name.
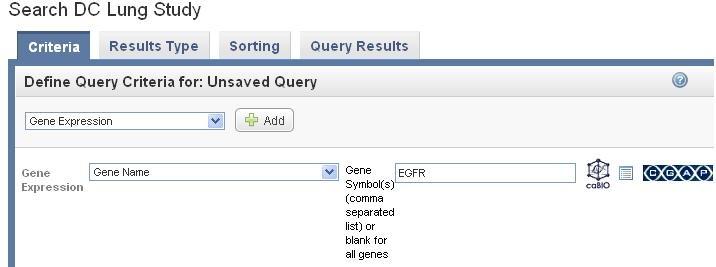
Step 4 Search for pathway in caBio.
Click on the caBIO icon at the right side to launch the search in caBio.
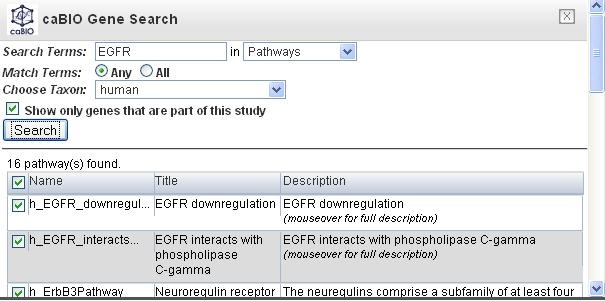
Put a search term in the field, and select pathways from the drop down menu. In this example, the search term EGFR was used and there are 16 pathways found in the result.
Step 5 Search for pathway gene.
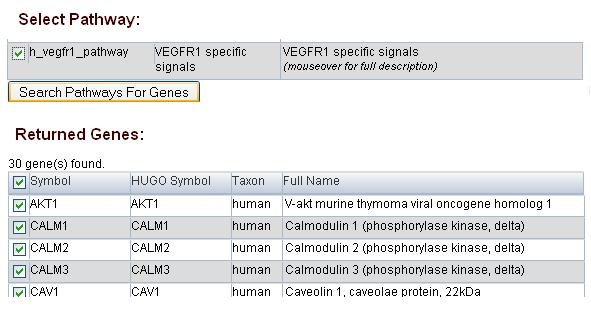
The user can select one or many pathways of interest to do the search, although the the list of genes returned will be large when the multiple pathways option is selected. In the example below, we selected one pathway : h_vegfr1_pathway (top portion of the figure) for the search and received 30 genes returned.
Step 6 Using pathway gene
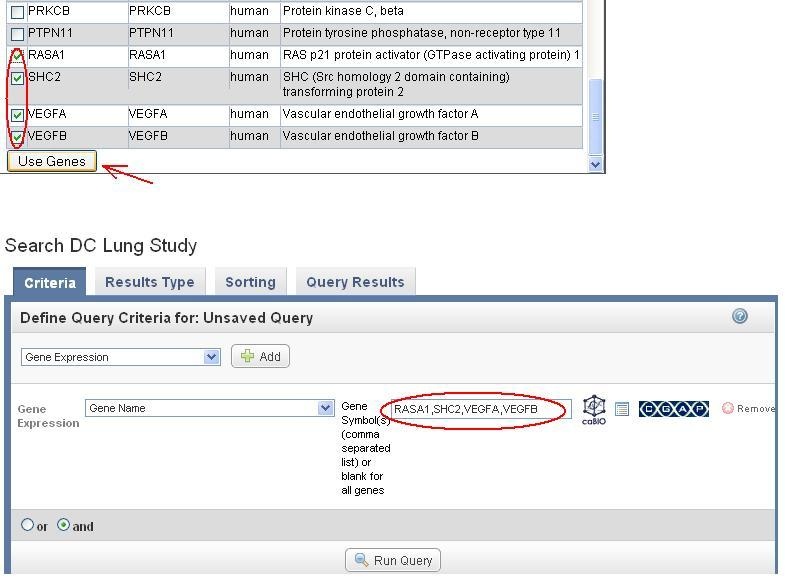
Select pathway genes of interest and click on "Use Genes" (top portion of the figure below). The selected genes are populated in the Gene Symbol box in the original Gene expression query (done at step 3) as shown below in the lower portion of the figure (RASA1, SHC2, VEGFA, VEGFP).
Have a comment?
Please leave your comment in the caIntegrator End User Forum.