Question: How can I build caArray from the source code in the NCI repository?
Topic: caArray Installation and Upgrade
Release: caArray v2.4.1.1 and higher
Date entered: 01/17/2012
Answer
Below is a step-by-step illustrated tutorial on how to build the application from source. It covers the following:
- How to check out the source code from the repository using a Subversion (SVN) client
- How to manually create the requisite database schema
- How to configure the various properties files required by Ant
- How to invoke the build process from the command line
- How to launch the caArray server once the build has completed successfully
- How to access the application's login page via your Web browser
The tutorial is designed for use in a Windows environment, but can easily be adapted to work in Linux as well.
Note
Due to recent changes in the NCI caArray source code repository, the process described in this tutorial only applies to v2.4.1.1, which is the latest version as of this writing.
Note
This tutorial is based on the readme.txt file in the NCI caArray source code repository at https://ncisvn.nci.nih.gov/svn/caarray2/tags/2.4.1.1/.
Prerequisites
1) Before proceeding to check out the code, ensure that the following are already installed on your machine:
- The Java 2 SE Development Kit (JDK) v1.5, available for download at:
http://download.oracle.com/otn/java/jdk/1.5.0_22/jdk-1_5_0_22-windows-i586-p.exe
Warning
The build process described in this tutorial only works with version 1.5 of the JDK; it will not work with any other versions, earlier or later.
- Apache Ant 1.7.0 or later, available for download at:
http://mirrors.ibiblio.org/apache//ant/binaries/apache-ant-1.8.3-bin.zip
- MySQL v5.0 or later, available for download at:
http://filehippo.com/download/file/3518c972e6317bff1a5caacae83b83cf1057fceca08180ebd44ad9a0ba01444b/
- A command-line Subversion (SVN) client of your choice. The one used in this tutorial is the Collabnet Subversion Command-Line Client v1.74, available for download at:
http://www.open.collab.net/servlets/OCNDirector?id=CSVN1.7.4WINC32 (registration required)
2) Once all the above are installed, ensure they are configured as follows:
- The PATH and JAVA_HOME Windows environment variables must be set to the the installation path of the JDK binaries. The default path in Windows is:
C:\Program Files\Java\jdk1.5.0_22\bin
- The PATH and ANT_HOME Windows environment variables must be set to the installation path of the Ant binaries. The default path is:
C:\ant
- The PATH Windows environment variable must be set to the installation path of the SVN client binaries. The default path in Windows is:
C:\Program Files\CollabNet Subversion Client
- The ANT_OPTS environment variable must be set to the value "-Xmx256m"
- The MySQL server must be configured with a secure password for the root user, and this password must be recorded for future reference
Checking Out The Source Code From The NCI Repository
To begin checking out the caArray source code from the NCI repository, first create a new folder (i.e., C:\source), then open up a command-line window, navigate to that folder, and enter the following:
svn checkout https://ncisvn.nci.nih.gov/svn/caarray2/tags/2.4.1.1/
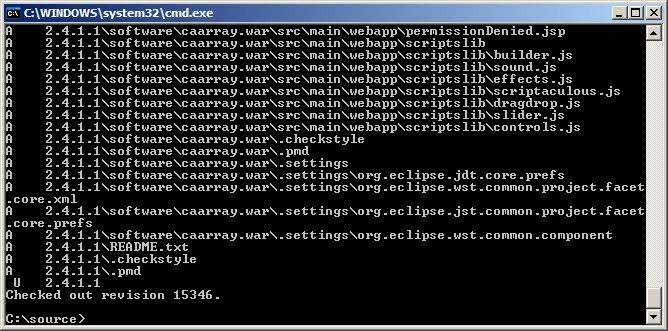
The source files will then begin downloading from the repository server into the folder you created. Depending on the speed of your Internet connection, it may take over half an hour for the checkout to complete, as the source contains thousands of files distributed over hundreds of folders.
You'll know when the checkout is complete when the command-line window shows a message stating, 'Checked out revision x', where x is the revision number, as shown in the screenshot below:
Manually Creating The Requisite Database Schema
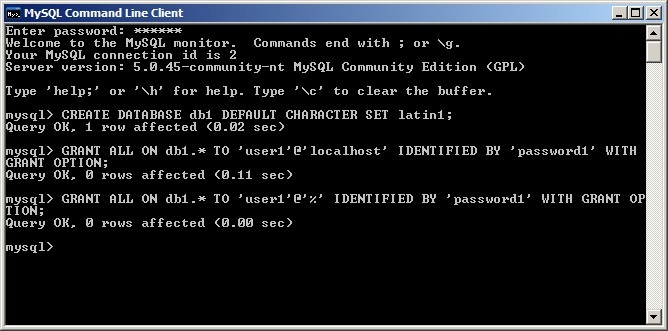
It is possible to have Ant automatically generate the empty database schema required for caArray to work. However, it is preferred to create these schema manually via the MySQL command line client. First, you need to derive a name for the database caArray will use to store genomic data (i.e., DB NAME), as well as a username (i.e., CAARRAY DB USER) and password (i.e., CAARRAY DB USER PASSWORD) for the user who is granted access to that database. In our example, the database name is db1, the username is user1, and the password name is password1.
Now, log into the console using the root password you set when installing the database server, then enter the following SQL commands line-by-line, substituting the database name, username, and password you derived where indicated:
CREATE DATABASE <DB NAME> DEFAULT CHARACTER SET latin1; GRANT ALL ON <DB NAME>.\* TO '<CAARRAY DB USER>'@'localhost' IDENTIFIED BY '<CAARRAY DB USER PASSWORD>' WITH GRANT OPTION; GRANT ALL ON <DB NAME>.\* TO '<CAARRAY DB USER>'@'%' IDENTIFIED BY '<CAARRAY DB USER PASSWORD>' WITH GRANT OPTION;
The client will respond with a confirmation that the issued queries were successful, as shown in the screenshot below:
Configuring The Various Properties Files Required By Ant
Invoking The Build Process From The Command Line
Launching The caArray Server Upon Build Completion
Accessing The Application's Login Page Via Your Web Browser
Have a comment?
Please leave your comment in the caArray End User Forum.