This appendix describes configurations for importing data into a study.
Topics in this appendix include the following:
Subject Annotation Data Configuration
The following subject annotation data configuration information is collected:
- Subject annotation Data Source (delimited text)
- Protocol Id (of study to import)
For delimited text, see Delimited-Text Annotation Import. For subject annotation files, one field must be identified as the subject identifier.
See Annotation Field Configuration for details on specification of visibility and browse configuration.
Delimited-Text Annotation Import
Delimited-text annotation files must be in standard comma-separated value format. The file must include a header line that specifies the name for each field. Each row of data must contain the same number of values as the header row. The file must include a column that will be designated as the identifier (for example, subject identifier, sample identifier, etc.) for each row. Each identifier must be unique. After upload of the file, the Study Manager must indicate for each field a Field Descriptor Type: Annotation or Identifier. For each Field Descriptor Type Annotation the Study Manager must select a Data Type: date, numeric or string.
After specification of these types, the file will be validated to ensure that the values are valid for the types selected and that the file conforms to the requirements given above.
For more information, see Working with Annotations.
Annotation Field Configuration
For each annotation field (regardless of the source), the Study Manager must specify the following information:
- Annotation semantics: each annotation field (whether associated with a subject, image series, image or sample) must either:
- be associated with an existing annotation definition known to the system,
- be associated to an existing CDE in caDSR or
- have sufficient semantic metadata recorded so that the field may be submitted for registration as a CDE in caDSR.
- Field authorization: Each field must be either declared publicly visible or restricted to a list of groups. The default will be the visibility settings given at the study level. For more information, see Define Fields Page for Editing Annotations.
- Whether the field is to be included in the results list for a given entity type (i.e. Subject, Sample, Image Series or Array Data) when browsing data.
- Whether the field is to be included in simple single-input searches when browsing data.
For more information, see Adding Subject Annotation Data.
Sample Data Configuration
Sample data can be uploaded from either caArray 2 or from delimited-text import. Samples imported from caArray 2 may have annotations updated by use of the delimited-text import functionality, if sample annotation is required. Import from caArray 2 requires specification of the following information:
- caArray server hostname
- caArray server JNDI port
- caArray username
- caArray password
- Either the experiment identifier (to import all samples in the experiment) or a file containing a comma-separated list of samples in the format "experiment identifier", "sample name".
- Mapping of samples to subjects. This may be specified by a comma-separated list in the format "subject identifier", "sample identifier" or by a regular-expression based mapping formula.
When samples are imported via delimited-text import, the time-point is associated to the sample itself. This means that each sample may be associated with only one time-point (i.e. multiple time-points for the same sample are invalid).
Genomic Data Configuration
All genomic data (i.e. array data) is imported from caArray 2. First the Study Manager must specify sufficient information to map study samples to caArray 2 samples. If all samples were imported directly from caArray 2 as described in Special Requirement: Sample Data Configuration, no further information is required for this step. If samples were imported via delimited-text, the Study Manager must specify
- caArray server hostname
- caArray server JNDI port
- caArray username
- caArray password
- A mapping of caIntegrator sample identifiers to caArray 2 samples, specified as a comma-separated list in the format "caIntegrator sample identifier", "caArray 2 experiment identifier", "caArray 2 sample name".
The system enables the Study Manager to navigate easily to the selected caArray 2 instance.
Next, the system indicates the available platforms and array data types available for the study samples. The Study Manager must indicate which platforms and data types to import and for each platform/data type combination must specify the following:
- Whether to import the data
- The visibility of the data, either public or restricted to a set of groups. Low-level genotyping data (raw data and normalized) always has restricted visibility.
See also Supplemental Files Configuration.
Supplemental Files Configuration
This section describes the format that must be used when creating supplemental files for use by caIntegrator. The supplemental files described here are to be added to an experiment in caArray prior to configuring a study in caIntegrator.
The file itself is a tab-delimited text file. The file extension can be anything, though users typically use .txt. The name of each supplemental file must be unique within a caArray experiment.
Inside the file, each row in the file contains the data from one reporter. Each column in the file must have a unique header name, that is, you cannot give two different columns the same column name.
There are two supported formats for these files: Single Sample Format and Multiple Sample Format.
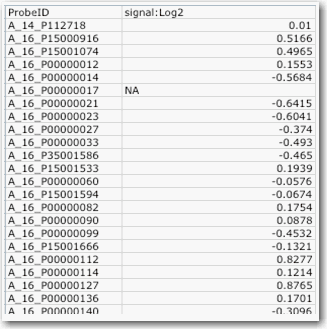
Single Sample Format
Single sample format requires the following parameters:
- Minimum of two required columns
- One column must contain the reporter/probe name
- One column must contain the value be reported by the reporter
- The file can have additional columns, though other than reporter/probe name and value mentioned above, the rest will be ignored
- One single sample file for each sample in the experiment
An example of single sample format file is shown in the following figure
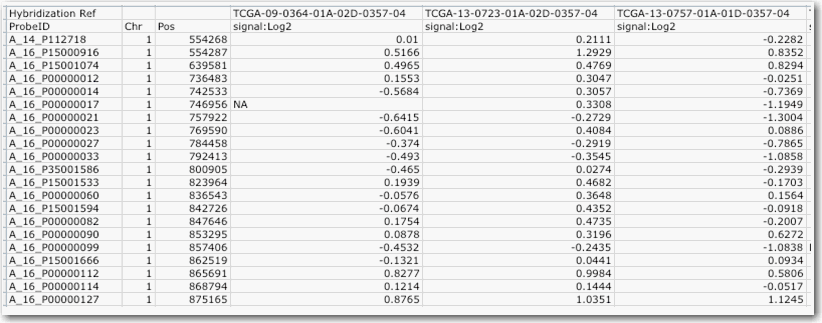
Multiple Sample Format
Multiple sample format requires the following parameters:
- One column must contain the reporter/probe name.
- Each additional columns are the reporter values such that there is one column per sample.
One multiple sample file for the whole experiment.
Note
Currently the multiple sample format is slower to load than the single sample format for platforms other than Agilent Copy Number. Future releases should show improvements in this performance.
An example of multiple sample format file is shown in the following figure.
The following software programs create the supplemental data format used by caArray:
- Affymetrix Expression Console – This software produces supplemental files. In Expression Console, use the "Export Result" function to create these files. Note that when you use an algorithm other than MAS5 to normalize the data (for example using RMA or Plier), Expression Console automatically creates a […
summary.txt] file that contains extra lines on top of the derived data results. The extra lines all start with a "#" to signify that it is a remark. These lines are ignored by caIntegrator parsing. - Agilent GeneSpring GX – This software can export a results table in
.txtformat.
Imaging Data Configuration
The following imaging data configuration information is collected:
- NBIA grid server hostname (defaults to CBIIT instance)
- NBIA grid server port (defaults to CBIIT instance port)
- Protocol Id
- Mapping of NBIA Patients to subjects imported from subject annotation data source. This may be specified by a comma-separated list in the format "subject identifier", "NBIA patient identifier" or by a regular-expression based mapping formula.
- Which annotation fields to import from NBIA.
The system enables the Study Manager to navigate easily to the selected caArray 2 instance.
Additional annotation for either images or image series can be imported using the delimited-text import functionality.